Fig. 3.
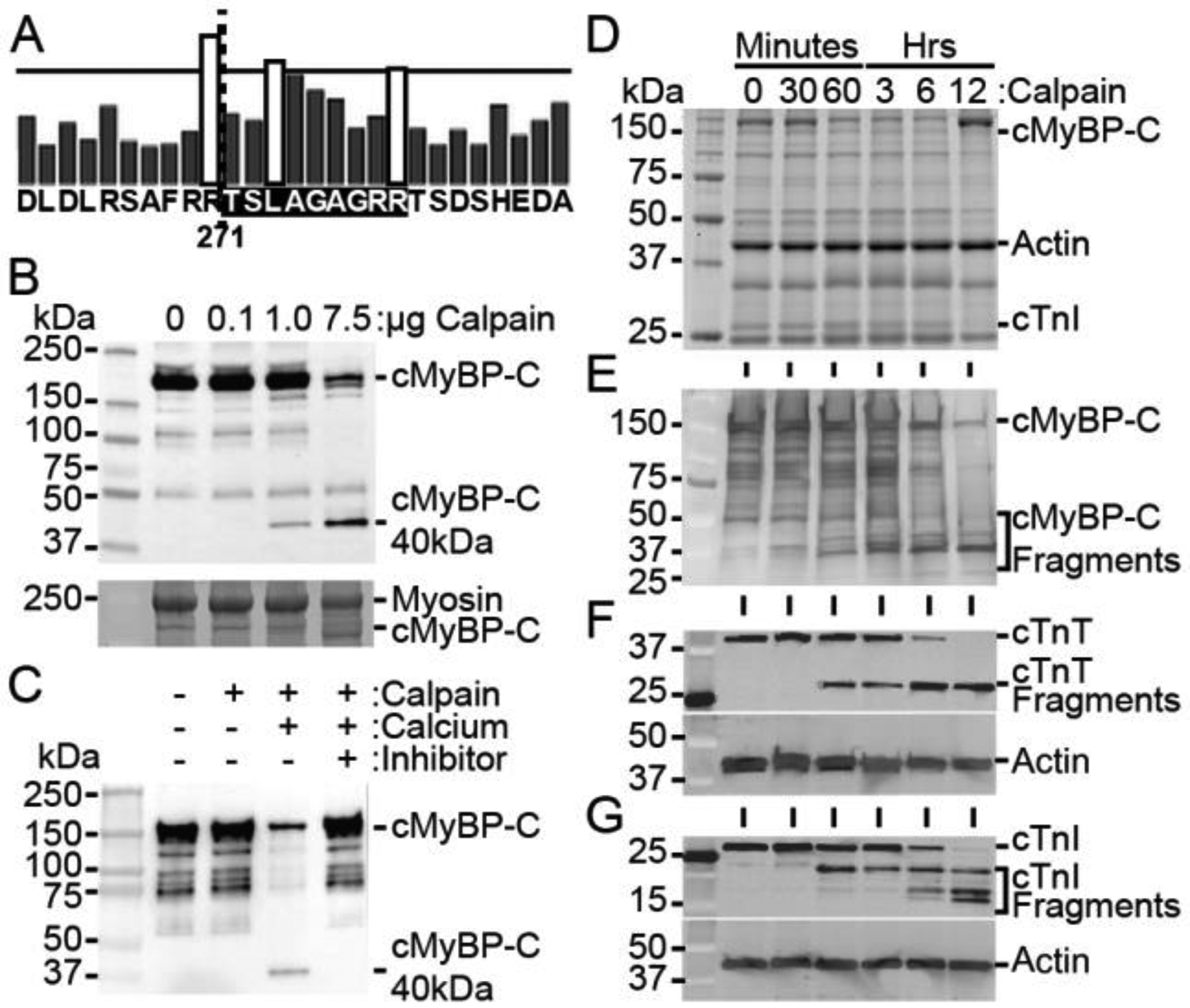
Calpain proteases degrade cMyBP-C and generate the 40kDa fragment. (A) In silico analysis of calpain proteolysis sites on cMyBP-C, with bar height representing the likelihood of cleavage, indicates R271 as a potential location of cleavage (dotted line), corresponding to the known sequence of the 40kDa fragment. The calpain-target site is highlighted in black. (B) Incubation of myofilaments with increasing concentration of calpain with 10 mM calcium for 1 hour at 37 °C shows a dose-dependent increase in cMyBP-C proteolysis and the generation of the 40kDa fragment with a SYPRO Ruby-stained SDS-PAGE loading control (bottom). (C) Proteolysis of cMyBP-C in myofilament fractions incubated with 1 μg calpain for 1 hour at 37 °C is prevented in the absence of calcium or in the presence of 10 nM of the calpain inhibitor MDL 28170. (D) Myofilament protein fractions demonstrate proteolysis at time points following incubation of 20 μg of total myofilament protein with 1 U μ-calpain with 10 mM calcium. (E) Western blotting for cMyBP-C shows reduction in full-length cMyBP-C and an increase in 40kDa cMyBP-C with longer calpain incubation. (F and G) The known calpain targets cTnT and cTnI show a reduction in full-length protein and an increase in degraded protein with increasing incubation time with calpain.
