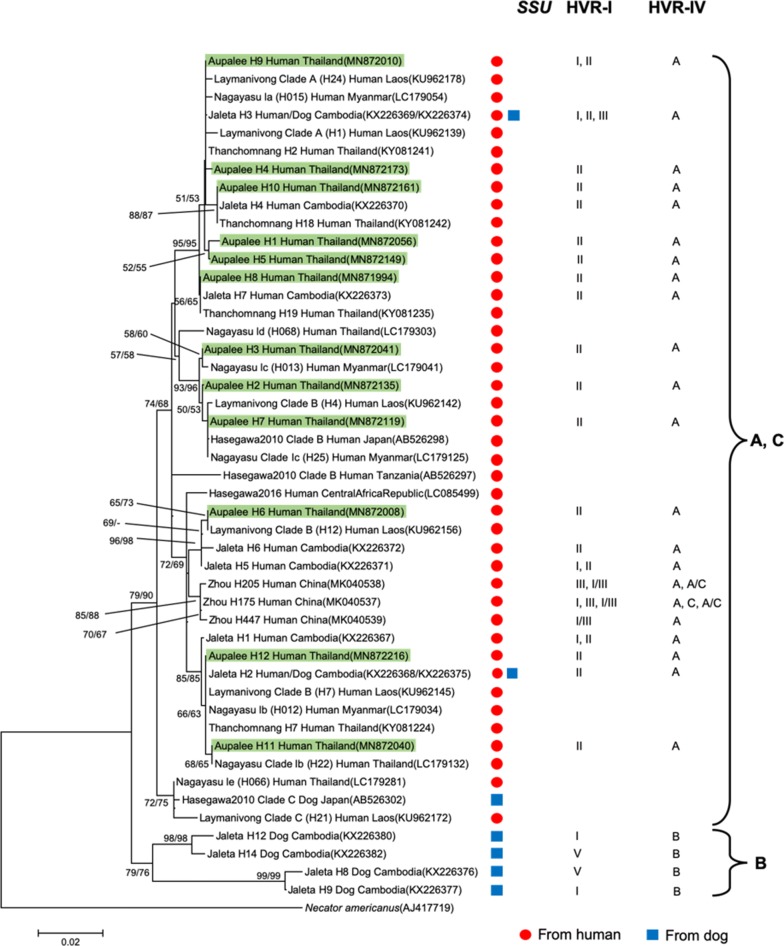
Fig. 1.
cox1 gene neighbor-joining tree of different S. stercoralis isolates based on 552-bp partial sequences. Shown are the sequences found in this study (green box) and selected published S. stercoralis haplotypes representing the major phylogenetic groups described in recent S. stercoralis cox1 phylogenies. The scale-bar represents 0.02 substitutions per site. The bootstrap values represent 1000 bootstrapping repetitions. Bootstrap values for neighbor-joining and maximum likelihood analysis are shown above or near the branches. The labels are composed as follows: author of the reference; haplotypes names according to this reference; host the isolate was derived from; country the isolate was isolated from; GenBank accession number. The host a particular sequence was found in is further highlighted with a red filled circle for “human” and a blue filled square for “dog”. The two columns on the right indicate the SSU HVR-I and HVR-IV haplotypes found among the worm individuals of the respective cox1 haplotype according to the respective authors, if known. Published sequences can be found under the following references [14–20]