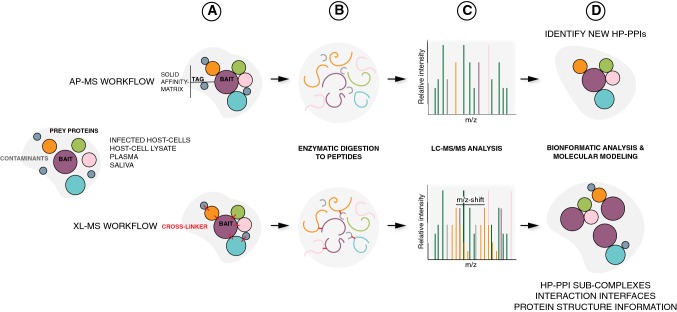
Fig. 2.
Schematic overview of the affinity-purification mass spectrometry (AP-MS) and cross-linking mass spectrometry (XL-MS) workflows. Interacting prey proteins (e.g., host proteins) to a given bait (e.g., bacterial protein) can be identified from a variety of biological mixtures, such as infected cells, host-cell lysates, plasma or saliva via AP-MS (top panel) or XL-MS (bottom panel). a In the AP-MS workflow, interacting prey proteins are enriched from the biological sample to an affinity-tagged bait protein attached to a solid affinity matrix; whereas in XL-MS, interacting prey proteins can be identified as associated to the bait via adding a suitable cross-linker to the sample and identifying cross-linked bait–prey peptides further down the workflow. b For the mass spectrometric identification of interacting proteins via either the AP-MS or the XL-MS workflow, all proteins present in either sample are digested to peptides via dedicated enzymes, prior to c mass spectrometric analysis of the samples via liquid chromatography tandem mass spectrometry (LC–MS/MS). In the XL-MS samples, a typical signature feature for a cross-linked peptide is an observable mass-over-charge (m/z) shift in the eluting peptides arising from isotopic variants of the cross-linker molecule. d Bioinformatic analysis of the acquired spectra allows for the identification of (novel) HP-PPIs and together with molecular modeling for the identification and structural determination of the HP-PPI sub-complexes and their interaction interfaces