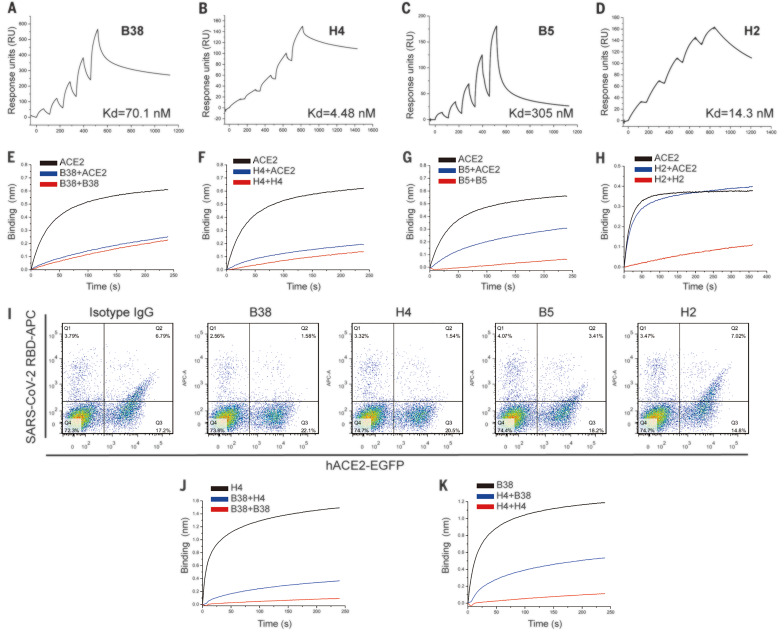
Fig. 1. Characterization of COVID-19 virus–specific neutralizing antibodies.
(A to D) The binding kinetics between four antibodies (B38, H4, B5, and H2) and COVID-19 virus RBD were measured using a single-cycle Biacore 8K system. (E to H) Competition binding to the COVID-19 virus RBD between antibodies and ACE2 was measured by BLI. Immobilized biotinylated COVID-19 virus RBD (10 μg/ml) was saturated with antibodies and then flowed with corresponding antibody in the presence of 300 nM soluble ACE2 (blue) or without ACE2 (red). As a control, the immobilized biotinylated RBD was flowed with buffer and then flowed with the equal molar concentration of ACE2 (black). The graphs show binding patterns after antibody saturation. (I) hACE2–enhanced green fluorescent protein (EGFP) was expressed on the HEK293T cell surface, and the cells were stained with 200 ng/ml COVID-19 virus RBD his-tag proteins preincubated with isotype IgG, B38, H4, B5, or H2. The percentages of anti-his-tag APC+ (allophycocyanin) cells and EGFP+ cells were calculated. (J and K) Competition binding to COVID-19 virus RBD between B38 and H4 was measured by BLI. Immobilized COVID-19 virus RBD (10 μg/ml) was saturated with 300 nM of the first antibody and then flowed with equal molar concentration of the first antibody in the presence of (blue) or without (red) the second antibody. Equal molar concentration of the second antibody was flowed on the immobilized RBD as a control (black). The graphs show binding patterns after saturation of the first antibody.