Abstract
The objective of this study was to compare the prevalence of human rotavirus group A common G and P genotypes in human Egyptian stool specimens and raw sewage samples to determine the most common genotypes for future vaccine development. From 1026 stool specimens of children with acute diarrhea and using nested RT-PCR, 250 samples (24.37%) were positive for human rotavirus group A. Using multiplex RT-PCR, rotavirus common P and G genotypes were detected as 89.20% and 46.40% of the positive clinical specimens respectively. This low percentage of common G genotypes frequency may affect the efficiency of the available live attenuated oral rotavirus vaccines [Rotarix® (human rotavirus G1P[8]) and RotaTeq® (reassortant bovine–human rotavirus G1-4P[5] and G6P[8])], however the percentage of clinical specimens which were negative for common G genotypes but positive for P[8] genotype was 12.00%. From 24 positive raw sewage samples for rotavirus group A VP6 collected from Zenin and El-Gabal El-Asfar wastewater treatment plants (WWTPs), 21 samples (87.50%) were typeable for common P genotypes while 13 samples (54.17%) were typeable for common G genotypes. Phylogenetic analysis of a VP8 partial gene of 45 P-typeable clinical isolates and 20 P-typeable raw sewage samples showed high similarity to reference strains and the majority of mutations were silent and showed lower to non-significant similarity with the two vaccine strains. This finding is useful for determining the most common antigens required for future vaccine development.
Electronic supplementary material
The online version of this article (10.1007/s12560-020-09426-0) contains supplementary material, which is available to authorized users.
Keywords: Human rotavirus group A, Surveillance, Common P and G genotypes, Diarrhea, Non-silent mutation
Introduction
Rotaviruses belong to the Reoviridae family and contain genomes consisting of eleven segments of dsRNA. The International Committee of Taxonomy of Viruses (ICTV) recognized eight species within the rotavirus genus: Rotavirus A, B, C, D, E. F, G, and H (Attoui et al. 2012). Viruses of the rotavirus group A (RVA) species had been the most widely studied, owing to their significance as the prime cause of severe diarrhea in infants and young children (Greenberg and Estes 2009). Rotaviruses group A were further classified into different G and P genotypes based on the molecular characterization of the outer capsid proteins, VP7 (glycoprotein) and VP4 (protease-sensitive, which was cleaved into VP5 and VP8 by trypsin) respectively (Matthijnssens et al. 2011). There are at least 51 P genotypes and 36 G genotypes of rotaviruses A recognized in humans and animals (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg) (accessed on Nov. 7, 2019). The most prevalent rotavirus genotypes detected around the world include four common human G genotypes (G1, G2, G3, G4) in association with the most common human P genotypes P[4], P[6], and P[8] (Bányai et al. 2012; Chen et al. 2012). The four common G genotypes (G1-G4) and the three common P genotypes (P[8], P[4] and P[6]) represented approximately 93% and 99% of the rotavirus reports in South Korea respectively (Than and Kim 2013). In Turkey, the four common G genotypes along with G9 accounted for 97.8% while the three common P genotypes represented more than 99% of rotavirus strains (Durmaz et al. 2014). The most common G/P genotypes combinations infecting humans worldwide were G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and G12P[8]. These genotypes were responsible for almost 90% of rotavirus infections worldwide (Matthijnssens and Van Ranst 2012; Li et al. 2016).
The World Health Organization (WHO), global rotavirus surveillance network estimated that the annual rotavirus-associated mortality is approximately 215,000 worldwide in children < 5 years of age (Tate et al. 2016). Currently, two live attenuated oral rotavirus vaccines, the monovalent Rotarix® (derived from a single strain of human rotavirus G1P[8]) and the pentavalent Rotateq® (containing five reassortant bovine–human rotavirus, G1-4P[5] and G6P[8]) had been licensed and used extensively in > 100 countries worldwide since 2006 (Parashar et al. 2016). However, these vaccines had been reported to be less efficient in African children and did not cover all circulating rotavirus G and P genotypes (Madhi et al. 2010; Harris et al. 2017).
In a previous study in Egypt, overall, 22 of 30 rotavirus-containing samples collected from Greater Cairo sewage in 1998–1999 could be P typed, and thus the percentage of untypeable samples was 26.67%. The frequencies of P genotypes were as follows: P[8], 53.33%; P[6], 30.00%; and P[4], 16.67%. It indicated the high frequency of the most common P genotypes (P[8], P[6], and P[4]) in the Egyptian community. While 21 of the 30 rotavirus-containing samples could be G typed, and thus the percentage of untypeable samples was 30.00% and the most frequent G genotype was G1 (69.60%), followed by G3 (13.00%), G4 (8.70%), and G9 (8.70%) (Villena et al. 2003; El-Senousy et al. 2004). Also, previous studies reported that rotavirus group A was the most frequent RNA enteric viruses in Egyptian clinical specimens and aquatic environment and was the most resistant one to sewage and water treatment processes (El-Senousy et al. 2004, 2013a, c, 2014a, b, c; El-Senousy and El-Mahdy 2009). Rotavirus group A was more frequent than rotavirus group C in Egyptian clinical specimens and environmental samples (El-Senousy et al. 2015).
Although, the WHO had recommended introduction of rotavirus vaccines into all national immunization program, more than 100 countries, including Egypt, had not introduced the vaccines to their compulsory immunization program (Karami and Berangi 2018). Thus, continuous surveillance of rotavirus genotypes is required to determine if the live attenuated oral vaccines could be used to cover the common genotypes circulating in the Egyptian infants. Furthermore, these vaccines had been associated with a low risk of intussusceptions which is believed to be triggered by the replication of the oral vaccines (Carlin et al. 2013; Weintraub et al. 2014; Yih et al. 2014; Yen et al. 2016). Also, these vaccines may be associated with other issues related to live vaccines including the risk of introduction of vaccine strains into the environment, genetic reassortment between the vaccine and a wild type strain, and reversion of the vaccine strain toward virulence (Lappalainen et al. 2015). This has stimulated interest in an alternative, non-living parenteral approach to immunization. Our group (Food-Borne Viruses Group, NRC) aimed to develop a parenteral, non-living recombinant subunit rotavirus vaccine and we need to determine the most common Egyptian antigens required for formulation of this vaccine. Thus, there is a need to determine the most common G and P genotypes circulating in Egyptian infants and children to suggest a vaccine that will cover the most circulating G and P genotypes in Egypt. The present study was conducted to estimate the burden of rotavirus gastroenteritis as well as to compare the prevalence of rotavirus common G and P genotypes among children ≤ 5 years of age visiting Abo El-Reech hospital (from Oct. 2015 to Sep. 2017) and in raw sewage samples collected from WWTPs in Greater Cairo during winter and autumn months in the same period to determine the most common G and P genotypes for future vaccine development. Additional objective was to estimate the silent and non-silent mutations in the human rotavirus VP8 partial gene in clinical specimens and raw sewage samples.
Materials and Methods
Collection of Clinical Specimens
One thousand and twenty six stool specimens were collected from Oct. 2015 to Sep. 2017 from children ≤ 5 years of age visiting Abo El-Reech hospital, in Greater Cairo, Egypt. The specimens included 651 males and 375 females. They were collected from children ≤ 5 years’ old suffering from acute diarrhea (Table 1). Specimens were collected in clean containers and transferred to the Virology Lab., Water Pollution Research Department, Environmental Research Division, NRC, within 3 h after collection for examination.
Table 1.
Number of clinical specimens collected from different age groups
| Age group (in months) | Number of collected specimens |
|---|---|
| ]0, 6] | 366 |
| ]6, 12] | 474 |
| ]12, 18] | 108 |
| ]18, 24] | 49 |
| ]24, 60] | 29 |
Concentration of Clinical Specimens
Approximately, 0.1 g of stool specimens was weighed, diluted 1:10 in nuclease-free water, and vortexed for 30 s. Specimens were centrifuged at 7000 rpm for 10 min at room temperature and the supernatant was kept in − 70 °C until used.
Viral Nucleic Acid Extraction
Viral nucleic acid was extracted from 100 µl of the supernatant by BIOZOL Total RNA Extraction reagent (BioFlux, Japan) according to the manufacturer’s instructions.
Nested RT-PCR of a VP6-Coding Gene Fragment of Rotaviruses Group A
The primers used for RT-PCR to amplify a 379-bp region were the forward VP6-F 5′-GACGGVGCRACTACATGGT-3′ and the reverse VP6-R 5′- GTCCAATTCATNCCTGGTGG-3′ primers (1 µM of each) and according to (Iturriza-Gomara et al. 2002) using 100 units of M-MLV reverse transcriptase enzyme (Thermo Fisher) in a total volume of 10 µl and 2.5 units of Taq DNA polymerase (Thermo Fisher) in a total volume of 50 µl. Nested PCR amplification of the target RT-PCR products was performed using the forward VP6-NF 5′-GCWAGAAATTTTGATACA-3′ and the reverse VP6-NR 5′-GATTCACAAACTGCAGA-3′primers 1 µM of each and according to (Gallimore et al. 2006) to amplify 155 bp fragment. Ten µl of PCR products were analyzed by 3% agarose gels (Panreac-Spain).
Raw Sewage Samples
Twenty four raw sewage samples (1 litre volume of each sample) were previously screened for rotavirus group A VP6 (Kamel et al. manuscript in preparation) and all of them were positive. These samples were collected from inlets of El-Gabal El-Asfar and Zenin WWTPs in autumn and winter months from Oct. 2015 to March 2016 and from Oct. 2016 to March 2017. Samples were concentrated using aluminum hydroxide precipitation method according to Standard Methods for the Examination of Water and Wastewater (APHA 2017). Viral nucleic acid was extracted by BIOZOL Total RNA Extraction reagent (BioFlux, Japan) according to the manufacturer's instructions. El-Gabal El-Asfar and Zenin WWTPs use an activated sludge as a treatment technology and the flow rate in El-Gabal El-Asfar was 1,700,000 cubic meters per day (m3/day) while the flow rate in Zenin was 330,000 m3/day. El-Gabal El-Asfar receives raw sewage of a large area in Cairo Governorate and Zenin receives raw sewage of a large area in El-Giza Governorate. Raw sewage of the two WWTPs can be used as an excellent model for the prevalence of common P and G genotypes in Greater Cairo.
Genotyping for Positive Human Rotavirus Group A VP6 Clinical Specimens and Raw Sewage Samples
The extracted dsRNA of the positive rotavirus VP6 stool specimens and raw sewage samples were analyzed for common G and P genotypes using the methods described by (Gouvea et al. 1990) and (Gentsch et al. 1992) respectively.
Human Rotavirus Group A Common G Genotyping Using Multiplex Nested RT-PCR
Human rotavirus group A G genotyping was performed using multiplex nested RT-PCR (Gouvea et al. 1990). The RT and the first round PCR amplified the whole gene segment 9 (coding for VP7) (1062 bp) using primers Beg9 5′-GGCTTTAAAAGAGAGAATTTCCGTCTGG-3′ and End9 5′- GGTCACATCATACAATTCTAATCTAAG-3′. The second round PCR was a multiplex nested PCR and included the primer RVG9 5′- GGTCACATCATACAATTCT-3′ and the G-type specific primers aBT1 (G1 specific) 5′-CAAGTACTCAAATCAATGATGG-3′, aCT2 (G2 specific) 5′-CAATGATATTAACACATTTTCTGTG-3′, aET3 (G3 specific) 5′-CGTTTGAAGAAGTTGCAACAG-3′, and aDT4 (G4 specific) 5′- CGTTTCTGGTGAGGAGTTG-3′, with a predicted product sizes of 749 bp, 652 bp, 374 bp, and 583 bp respectively.
The PCR mixture for the first round consisted of 5 µl of the PCR buffer (Roche), 0.2 mM of each dNTP’s, 1 U of Expand PCR enzyme (Roche) and 100 µM concentration of each primer. The first round PCR was performed by adding 5 µl of cDNA [primed with 1 µM of both Beg 9 and End 9, 0.2 mM of dNTP's and 3 U of RT enzyme (Roche); RT was done at 50 °C for 1 h] to 45 µl of PCR mixture. After denaturation at 95 °C for 9 min, 40 PCR cycles which each consisted of 94 °C for 1 min, 47 °C for 2 min, and 72 °C for 5 min were performed, followed by an extension at 72 °C for 10 min. The second round PCR was performed using 2 µl of the first round reaction product in the same mixture described above but involving all the multiplex primers. The PCR protocol was as follows, denaturation at 95 °C for 9 min, 35 PCR cycles which each consisted of 94 °C for 1 min, 45 °C for 2 min, and 72 °C for 3 min followed by an extension at 72 °C for 10 min. Ten µl of PCR products were analyzed by 3% agarose gels (Panreac-Spain).
Human Rotavirus Group A Common P Genotyping Using Multiplex Semi-nested RT-PCR
Human rotavirus group A P genotyping was performed using multiplex semi-nested RT-PCR (Gentsch et al. 1992). The RT and the first round PCR amplified the VP8 fragment (876 bp) of the gene segment 4 (coding for VP4) using primers Con2 5′- ATTTCGGACCATTTATAACC-3′ and Con3 5′-TGGCTTCGCCATTTTATAGACA-3′. The second round PCR included the primers Con3 and the P-type specific primers 1T-1 (P[8] specific) 5′-TCTACTTGGATAACGTGC-3′, 2T-1 (P[4] specific) 5′-CTATTGTTAGAGGTTAGAGTC-3′, and 3T-1 (P[6] specific) 5′-TGTTGATTAGTTGGATTCAA-3′, with a predicted product sizes of 346 bp, 483 bp, and 267 bp respectively.
The RT reaction with primers Con2 and Con3 was performed in a similar way that in G typing. The PCR mix for the first and second rounds of amplification was the same as that for the G typing except for annealing temperature of the second round PCR that was 44 °C. Ten µl of PCR products were analyzed by 3% agarose gels (Panreac-Spain).
Monoplex Nested RT-PCR for Uncommon G Genotyping
It was performed like the multiplex nested RT-PCR except for using the G-type specific primers separately in the second round PCR. These primers were aFT5 primer (G5 specific) 5′-GACGTAACAACGAGTACATG-3′, aT8 (G8 specific) 5′-GTCACACCATTTGTAAATTCG-3′, and aFT9 (G9 specific) 5′-CTAGATGTAACTACAACTAC-3′ with a predicted product sizes of 303 bp, 885 bp, and 306 bp respectively according to (Gouvea et al. 1990, 1994; Villena et al. 2003).
Monoplex Semi-nested RT-PCR for Uncommon P Genotyping
It was performed like the multiplex semi-nested RT-PCR except for using the P-type specific primers separately in the second round PCR. The primer used was 4T-1 (P[9] specific) 5′-TGAGACATGCAATTGGAC-3′, with a predicted product size of 391 bp according to (Gentsch et al. 1992; Villena et al. 2003).
Statistical Analysis
For gender rotavirus infection, rotavirus positive cases were compared with rotavirus negative cases, using Person Chi-square (χ2) test while differences between percentages of rotavirus common P and G genotypes were compared using McNemar’s test to determine statistical significance. P value < 0.05 was considered statistically significant. Confidence intervals (CI) for all calculations were set at 95%.
Amplification, Sequencing, and Phylogenetic Analysis of Human Rotavirus VP8 Partial Fragment of the VP4 of Clinical Specimens and Raw Sewage Samples
RNA extracts of 45 from the positive common P-typeable clinical specimens in addition to 20 positive common P-typeable raw sewage samples were selected for amplification of a partial VP8 fragment of the VP4 positive gene according to (Wen et al. 2012). The selected positive clinical specimens RNA extracts included 17 specimens contained P[8] genotype (ID: EGY/SS1 to SS17), 18 specimens contained P[4] genotype (ID: EGY/SS18 to SS35) and 10 clinical specimens with P[6] genotype (ID: EGY/SS36 to SS45). The positive common P-typeable raw sewage samples included 7 samples contained P[8] genotype (EGY/RSS1 to RSS7), 9 samples contained P[4] genotype (EGY/RSS8 to RSS16), and 4 samples contained P[6] genotype (EGY/RSS17 to RSS20). The primers used for constructing VP8 amplified fragment were as follows 5′-TACTCATATGTTAGATGGTCCTTATCAGCCAAC-3′ (Wa ΔVP8* sense), 5′-TAGAGCTCTATCACAGACCATTATTAATATATTCATTAC-3′ (Wa ΔVP8* antisense) to amplify 477 bp (nt 202 to nt 678) fragment of P[8] VP8, 5′-TACTCATATGGTTTTAGATGGTCCTTATCAAC-3′ (DS-1 ΔVP8* sense) and 5;- TAGAGCTCTATCATAAACCATTATTGATATACTCG -3′ (DS-1 ΔVP8* antisense) to amplify 480 bp (nt 190 to nt 669) fragment of P[4] VP8, and 5′-TACTCATATGGTACTCGATGGTCCTTATCAACC-3′ (1076 ΔVP8* sense) and 5′ TAGAGCTCTATCATAACCCAGTATTTATATATTCATTACAC-3′ (1076 ΔVP8* antisense) to amplify 480 bp (nt 199 to nt 678) fragment of P[6] VP8. These amplified fragments encode amino acid residues 65–223 of Wa VP8*, 64–223 of DS-1 VP8*, and 64–223 of 1076 VP8*. VP8* cDNA was synthesized by using Superscript III (Invitrogen) and reaction parameters were as follows: 50–200 ng of genomic RNA was denatured with a final concentration of 15% DMSO and incubated at 94 °C for 3 min, followed by chilling immediately. The first strand cDNA was synthesized following manufacturer's instructions. The products of RT reaction were used as templates for PCR using an iProof High-Fidelity PCR system (Bio-Rad) to amplify the truncated VP8* fragments of rotaviruses with P[8], P[4], and P[6] specificity. The PCR products of positive VP8* fragments of rotaviruses P[8], P[4], and P[6] were sequenced. Fifty to one hundred µl of the PCR products were purified using a high pure PCR products purification kit (Qiagen) and following the manufacturer's instructions. Sequencing was performed on 1–7 µl of the purified products with an API prism Big dye termination cycle sequencing ready reaction kit (Applied Biosystem) using the same primers as in the PCR and following the manufacturer's instructions. The DNA was sequenced with an API prism 310 automated DNA sequencer. Sequence analysis was conducted to examine the silent and non-silent mutations in the amplified truncated VP8* fragments. Sequence analysis was conducted also to examine the phylogenetic and the evolutionary relationship between the Egyptian common P-typeable isolates and the vaccine strains of both Rotarix® and RotaTeq® vaccines and with some reference strains recorded in the GenBank. Strains were determined by BLAST (Basic Local Alignment Search tool) accessible at https://blast.ncbi.nlm.nih.gov/Blast.cgi from National Centre for Biotechnology Information (NCBI). Sequences were aligned along with other reference rotavirus sequences retrieved from GenBank using CLUSTAL W program in Mega X software (https://www.megasoftware.net). A neighbor joining tree was constructed with 1000 bootstrap replicates using Mega X software (https://www.megasoftware.net).
Results
Molecular Detection of a VP6-Coding Gene Fragment of Rotavirus Group A
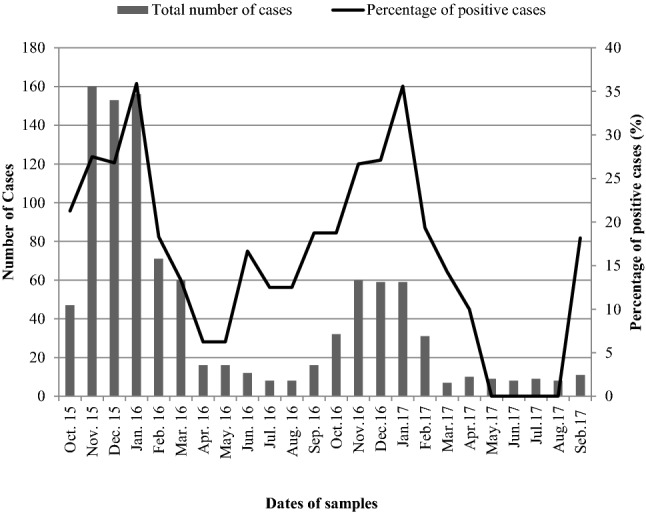
A total of 250 clinical specimens out of 1026 (24.37%) collected from October 2015 to September 2017 from Abo El-Reech hospital were positive for rotavirus VP6. The seasonal frequency of rotavirus detection was highest during November to January. The highest peak for rotavirus detection was in January 2016 (35.90%) and January 2017 (35.59%) (Fig. 1).
Fig. 1.
Seasonal variations of rotavirus group A in Egyptian cases
Rotavirus A was detected in 26.27% (171/651) males and 21.07% (79/375) females. Although these results seemed to indicate that male is the more infected gender, the statistical analysis using the Person Chi-square test indicated that no statistically significant differences could be established (P value was 0.0617).
Rotavirus incidence was low in children ≤ 6 months [18.31% (67/366)] of age. The incidence increased in the age group > 6 and ≤ 12 months [30.59% (145/474)] followed by slight decrease in rotavirus infection in children aged > 12 and ≤ 18 months [27.78% (30/108)]. Then rapid decrease in the rotavirus incidence was noted with increasing age to 24 months [16.33% (8/49)]. No rotavirus infections were observed in children older than 24 months (Fig. 2).
Fig. 2.

Distribution of rotavirus group A in different ages
Distribution of Rotavirus Common and Uncommon G and P Genotypes Among Rotavirus VP6 Positive Clinical Specimens
Of 250 positive VP6 cases and using multiplex nested RT-PCR, 116 (46.40%) were typeable while 134 (53.60%) were untypeable for common G genotypes. Furthermore, of the 250 VP6 positive cases and using multiplex semi-nested RT-PCR, 223 (89.20%) were typeable, while 27 (10.80%) were untypeable for common P genotypes. Moreover, a total of 94 out of 250 (37.60%) of rotavirus VP6 positive cases could be typed for both common G and P genotypes, while 5 out of 250 (2.00%) of specimens could not be typed for either common G or P genotypes. A total of 129 out of 250 (51.60%) of rotavirus VP6 positive cases were typeable for common P but untypeable for common G genotypes, while 22 out of 250 (8.80%) were typeable for common G but untypeable for common P genotypes. The results seemed to indicate higher prevalence of common P genotypes than common G genotypes in the rotavirus positive specimens. The statistical analysis using McNemar’s test indicated that there was extremely significant difference [the odds ratio was 0.171 (CI 0.103 to 0.269) and the two-tailed P value was less than 0.0001] for distribution of rotavirus common P and common G genotypes in the rotavirus positive children.
Remarkably and using multiplex nested RT-PCR, the most prevalent G genotype in the rotavirus VP6 positive specimens was G1 (65/250, 26.00%) followed by G3 (51/250, 20.40%). There were not any specimens contained more than one G genotype. Genotypes G2 and G4 were absent in the present study. Using monoplex nested RT-PCR, uncommon G9 was detected in 31 specimens of the 250 rotavirus VP6 positive specimens (12.40%).There were not any specimens contained G9 with other common G genotypes. The G-untypeable specimens were 103/250 (41.20%) (Fig. 3). Likewise and using multiplex semi-nested RT-PCR, the most prevalent P genotype in the rotavirus VP6 positive specimens was P[4] (100/250, 40.00%) followed by P[8] (57/250, 22.80%) and finally P[6] (49/250, 19.60%). Using monoplex semi-nested RT-PCR, genotype P[9] was not detected in any of the rotavirus VP6 positive specimens. Mixed P genotypes were detected in 17/250 specimens (6.80%) which included 8 cases (8/250, 3.20%) contained P[8] genotype mixed with P[6] genotype, 6 cases (6/250, 2.40%) contained P[8] genotype mixed with P[4] genotype, and 3 cases (3/250, 1.20%) contained P[8] genotype mixed with both P[4] and P[6] genotypes (Fig. 4).
Fig. 3.
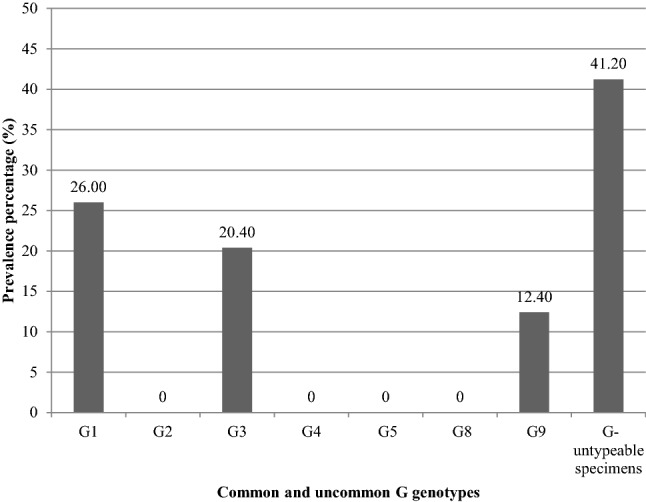
Frequency of common and uncommon G genotypes in the total positive rotavirus VP6 clinical specimens
Fig. 4.
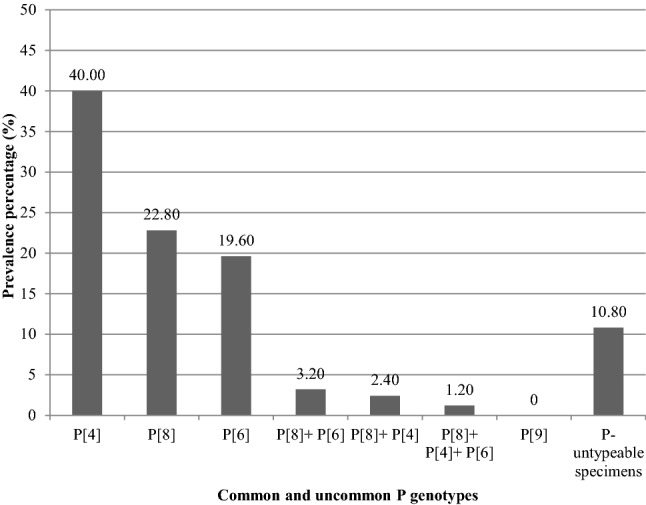
Frequency of common and uncommon P genotypes in the total positive rotavirus VP6 clinical specimens
Distribution of Rotavirus Common G Genotypes and either P[8], P[4] or P[6] Genotype Among Rotavirus VP6 Positive Clinical Specimens
The percentage of the common G-typeable cases [46.40% (116/250)] in addition to the percentage of the common G-untypeable cases but typeable for P[4] genotype [22.00% (55/250)] was 68.40% (171/250). Furthermore, the percentage of the common G-typeable cases (46.40%) in addition to the percentage of the common G-untypeable cases but typeable for P[6] [12.80% (32/250)] was 59.20% (148/250). Moreover, the percentage of the common G-typeable cases (46.40%) in addition to the percentage of the common G-untypeable cases but typeable for P[8] [12% (30/250)] was 58.40% (146/250).
Distribution of Rotavirus Common P Genotypes and either G1 or G3 Genotype Among Rotavirus VP6 Positive Clinical Specimens
The percentage of the common P-typeable cases [89.20% (223/250)] in addition to the percentage of the common P-untypeable cases but typeable for G1 [5.60% (14/250)] was 94.80% (237/250). Furthermore, the percentage of the common P-typeable cases (89.20%) in addition to the percentage of the common P-untypeable cases but typeable for G3 [3.20% (8/250)] was 92.40% (231/250).
Distribution of Rotavirus Common and Uncommon G and P Genotypes in Raw Sewage Samples
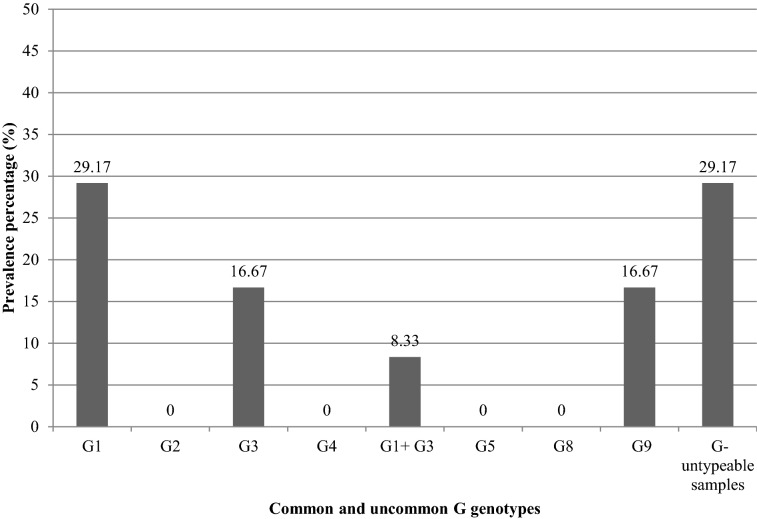
Of 24 positive VP6 raw sewage samples, 13 (54.17%) (7 raw sewage of El-Gabal El-Asfar and 6 raw sewage of Zenin) were typeable for common G genotypes. Furthermore, 21 samples (87.50%) (11 raw sewage of El-Gabal El-Asfar and 10 raw sewage of Zenin) were typeable for common P genotypes. Twelve samples (50%) (6 raw sewage of El-Gabal El-Asfar and 6 raw sewage of Zenin) could be typed for both common G and P genotypes. Nine samples (37.5%) (5 raw sewage of El-Gabal El-Asfar and 4 raw sewage of Zenin) were typeable for common P genotypes but untypeable for common G genotypes. One sample (4.17%) (raw sewage of El-Gabal El-Asfar) was typeable for common G genotypes but untypeable for common P genotypes. Two raw sewage samples of Zenin (8.33%) were untypeable for common G and P genotypes. The results again seemed to indicate higher prevalence of common P genotypes than common G genotypes in the rotavirus positive raw sewage samples with extremely significant difference as indicated by the statistical analysis using McNemar’s test [the odds ratio was 0.111 (CI 0.003 to 0.802) and the two-tailed P value equals 0.0269].
Using multiplex nested RT-PCR, the most prevalent G genotype in the positive rotavirus VP6 raw sewage samples was G1 (9/24, 37.50%) (5 raw sewage of El-Gabal El-Asfar and 4 raw sewage of Zenin) which 7 samples (7/24, 29.17%) (4 raw sewage of El-Gabal El-Asfar and 3 raw sewage of Zenin) contained G1 genotype without mixing with other common G genotypes and two samples (2/24, 8.33%) (one raw sewage of El-Gabal El-Asfar and one raw sewage of Zenin) contained G1 genotype mixed with G3 genotype. Second to G1 genotype, G3 genotype was detected in 6 samples (6/24, 25.00%) (3 raw sewage of El-Gabal El-Asfar and 3 raw sewage of Zenin) which 4 samples (4/24, 16.67%) (2 raw sewage of El-Gabal El-Asfar and 2 raw sewage of Zenin) contained G3 genotype without mixing with other common G genotypes and two samples (2/24, 8.33%) (1 raw sewage of El-Gabal El-Asfar and 1 raw sewage of Zenin) contained G3 genotype mixed with G1 genotype. G2 and G4 genotypes were absent in the present study (Fig. 5). Likewise and using multiplex semi-nested RT-PCR, the most prevalent P genotype in the positive rotavirus VP6 raw sewage samples was P[4] (10/24, 41.67%) (6 raw sewage of El-Gabal El-Asfar and 4 raw sewage of Zenin) which 9 samples (9/24, 37.50%) (5 raw sewage of El-Gabal El-Asfar and 4 raw sewage of Zenin) contained P[4] genotype without mixing with other common P genotypes and one sample (1/24, 4.17%) (raw sewage of El-Gabal El-Asfar) contained P[4] genotype mixed with P[8] genotype. Second to P[4] genotype, P[8] genotype was detected in 8 samples (8/24, 33.33%) (4 raw sewage of El-Gabal El-Asfar and 4 raw sewage of Zenin) which 7 samples (7/24, 29.17%) (4 raw sewage of Zenin and 3 raw sewage of El-Gabal El-Asfar) contained P[8] genotype without mixing with other common P genotypes and one sample (1/24, 4.17%) (raw sewage of El-Gabal El-Asfar) contained P[8] genotype mixed with P[4] genotype. Finally, P[6] genotype was detected in 4 samples (4/24, 16.67%) (two raw sewage of El-Gabal El-Asfar and two raw sewage of Zenin) which all of them contained P[6] genotype without mixing with P[4] and/or P[8] genotype (Fig. 6).
Fig. 5.
Frequency of common and uncommon G genotypes in the total positive rotavirus VP6 raw sewage samples
Fig. 6.
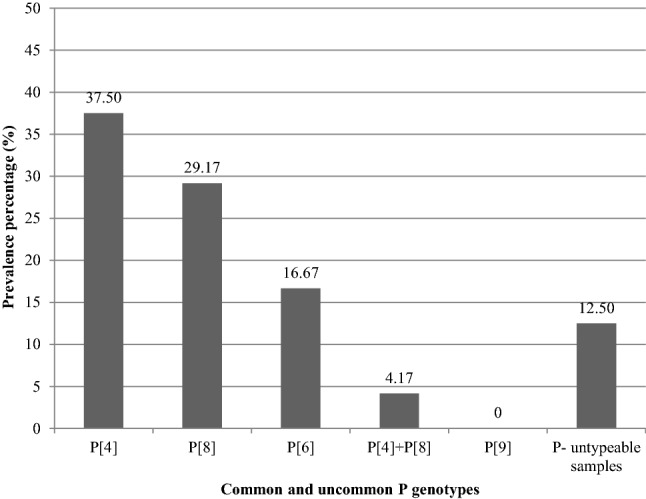
Frequency of common and uncommon P genotypes in the total positive rotavirus VP6 raw sewage samples
Using monoplex nested RT-PCR, uncommon G genotype G9 was detected in 16.67% (4/24) (two raw sewage of El-Gabal El-Asfar and two raw sewage of Zenin) of the positive rotavirus VP6 raw sewage samples (Fig. 5). All these positive samples were detected in the untypeable samples for common G genotypes in the multiplex nested RT-PCR. Uncommon genotypes G5 and G8 were completely absent in the present study (Fig. 5). Uncommon P[9] genotype was completely absent in this study (Fig. 6).
Sequencing of Human Rotavirus VP8 Partial Gene
Sequence analysis of human rotavirus VP8 gene from 17 clinical isolates and 7 raw sewage samples which contained P[8] genotype (Tables S1 and S2) showed highest relation to the two human rotavirus reference strains, Wa strain and rotavirus A strain USA (GenBank nucleotide accessions: FJ423116.1 and Kj659467.1, GenBank protein accessions: ACR22783.1 and AHW80479.1 respectively) (Fig. 7) with 97.06–99.16% nucleotide identity and 95.60–100% amino acid identity (Table 2). Of the seventeen P[8] VP8 aligned sequenced clinical isolates, 3 isolates collected during Oct. 2015, Nov. 2015, and Oct. 2016 (ID: EGY/SS1, SS3, and SS10 respectively) and one raw sewage sample [Zenin Jan. 2016 (ID: EGY/RSS3)] had similar sequences and showed four nucleotide substitutions with 99.16% nucleotide identity to the two human rotavirus reference strains. The other 12 clinical isolates [1 collected during Nov. 2015, 3 during Dec. 2015, 2 during Jan. 2016, 1 during Oct. 2016, 1 during Nov. 2016, 2 during Dec. 2016, and 2 during Jan. 2017 (ID: EGY/SS2, SS4, SS5, SS6, SS7, SS8, SS11, SS13, SS14, SS15, SS16, and SS17 respectively)] and 5 raw sewage samples [Zenin Nov. 2015, El-Gabal El-Asfar Dec.2015, El-Gabal El-Asfar March 2016, Zenin Oct. 2016, and Zenin Feb. 2017 (ID: EGY/RSS1, RSS2, RSS5, RSS6, and RSS7 respectively)] had similar sequences and showed 7 nucleotide substitutions with 98.53% nucleotide identity. All of them showed 100% homology in their amino acid sequences in comparison to the two human rotavirus reference strains (Table 2). Only two isolates of the 17 sequenced P[8] clinical specimens collected during Jan. 2016 and Nov. 2016 (EGY/SS9 and SS12 respectively), and one raw sewage sample [El-Gabal El-Asfar Jan. 2016 (ID: EGY/RSS4)] had similar sequences and showed 14 nucleotide substitutions in the coding sequence with 7 amino acid changes (i.e. non-silent mutations: Pro to Arg, Asn to Lys, Arg to Ser, Trp to Cys, Asn to Lys, Leu to Pro, and Asn to Lys at positions 76, 78, 117, 137, 149, 163, and 195 respectively). They showed 97.06% nucleotide identity and 95.60% amino acid identity in comparison to the two human rotavirus reference strains (Table 2).
Fig. 7.
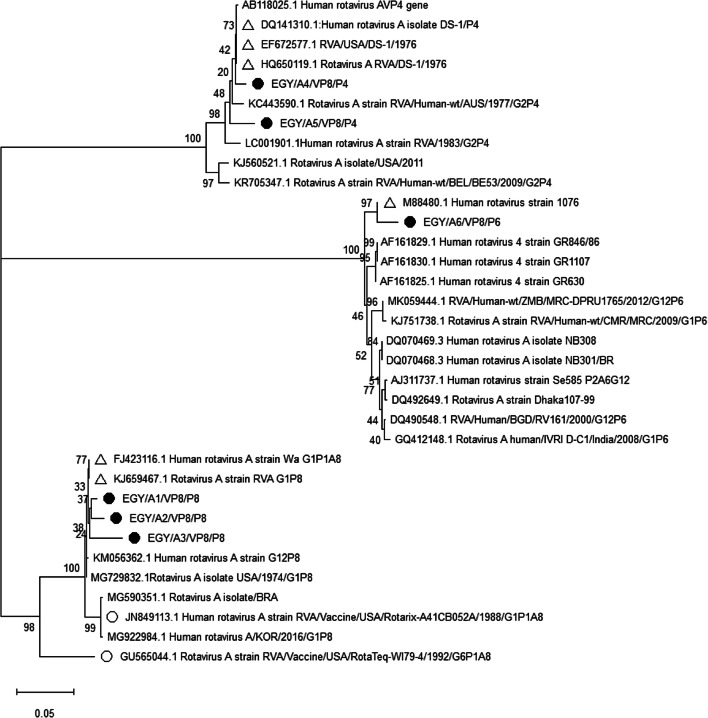
Phylogenetic tree of partial VP8 gene sequences of group A rotaviruses from P[8], P4], and P[6]. The evolutionary history was inferred using the Neighbor-Joining method. The sequenced studied Egyptian clinical specimens (EGY/SS1-EGY/SS45) and raw sewage samples (EGY/RSS1-EGY/RSS20) are shown with filled circles with ID from EGY/A1 to EGY/A6. EGY/A1 represented 4 isolates (EGY/SS1, SS3, SS10, and RSS3). EGY/A2 represented 17 isolates (EGY/SS2, SS4-SS8, SS11, SS13-SS17, RSS1, RSS2, and RSS5-RSS7). EGY/A3 represented 3 isolates (EGY/SS9, SS12, and RSS4). EGY/A4 represented 23 isolates (EGY/SS18-SS20, SS22, SS24-SS32, SS34, SS35, RSS8-RSS12, and RSS14-RSS16). EGY/A5 represented 4 isolates (EGY/SS21, SS23, SS33, and RSS13). EGY/A6 represented 14 isolates (EGY/SS36-SS45, RSS17-RSS20). The reference strains that showed highest similarity are labeled with empty triangles while the vaccine strains are labeled with empty circles and ID started with their GenBank accession numbers
Table 2.
Comparison the similarities of nucleotide sequences and amino acids sequences between Egyptian rotavirus A VP8 P[8] partial gene (from Egyptian infants and raw sewage samples) and RVA reference strains P[8] sequences as well as the representative Rotarix® and Rotateq® strains
| Strain name | Genotype | Nucleotide accession | Protein accession | Nucleotide identity with the Egyptian sequences | Amino acids identity with the Egyptian sequences | |||
|---|---|---|---|---|---|---|---|---|
| EGY/SS1, SS3, SS10, RSS3 (%) | EGY/SS2, SS4–SS8, SS11, SS13–SS17, RSS1, RSS2, RSS5-RSS7 (%) | EGY/SS9, SS12, RSS4 (%) | EGY/SS1–SS8, SS10–SS11, SS13–SS17, RSS1–RSS3, RSS5–RSS7 (%) | EGY/SS9, SS12, RSS4 (%) | ||||
| Rotavirus A strain/Wa | G1P1A[8] | FJ423116.1 | ACR22783.1 | 99.16 | 98.53 | 97.06 | 100 | 95.60 |
| Rotavirus A strain USA | G1P[8] | Kj659467.1 | AHW80479.1 | 99.16 | 98.53 | 97.06 | 100 | 95.60 |
| RVA vaccine/USA/Rotarix® | G1P1A[8] | JN849113.1 | AEX30660.1 | 97.48 | 96.86 | 95.39 | 98.11 | 93.71 |
| RVA vaccine/USA/Rotateq® | G6P1A[8] | GU565044.1 | ADK26989.1 | 90.93 | 90.72 | 89.03 | 94.34 | 91.19 |
Alignment of the nucleotide sequences and amino acid sequences of the P[8] VP8 sequenced Egyptian clinical isolates and raw sewage samples with the vaccine strain Rotarix® (GenBank nucleotide and protein accessions: JN849113.1 and AEX30660.1 respectively) showed 95.39–97.48% nucleotide identity and 93.71–98.11% amino acid identity (Table 2). Of the 17 P[8] VP8 sequenced Egyptian clinical isolates, 2 isolates collected during Jan. 2016 and Nov. 2016 (EGY/SS9 and SS12 respectively) and one raw sewage sample [El-Gabal El-Asfar Jan. 2016 (ID: EGY/RSS4)] showed 10 amino acid changes: Pro to Arg, Asn to Lys, Arg to Ser, Met to Thr, Ser to Arg, Trp to Cys, Asn to Lys, Leu to Pro, Phe to Leu, and Asn to Lys at positions 76, 78, 117, 120, 131, 137, 149, 163, 167, and 195 receptively. The remaining 15 clinical isolates and the 6 raw sewage samples showed 3 of the same amino acid changes at positions: 120, 131 and 167.
Alignment of the nucleotide sequences and amino acid sequences of the P[8] VP8 sequenced Egyptian clinical isolates and raw sewage samples with the vaccine strain Rotateq® (GenBank nucleotide accession: GU565044.1, and GenBank protein accession: ADK26989.1) showed 89.03–90.93% nucleotide identity and 91.19–94.34% amino acid identity. Of the 17 P[8] VP8 sequenced Egyptian clinical isolates, 2 clinical isolates collected during Jan. 2016 and Nov. 2016 (EGY/SS9 and SS12 respectively) and one raw sewage sample [El-Gabal El-Asfar Jan. 2016 (ID: EGY/RSS4)] showed 14 amino acid changes: Pro to Arg, Ile to Lys, Arg to Ser, Val to Ile, Asn to Ser, Asp to Asn, Trp to Cys, Asn to Lys, Lys to Arg, Leu to Pro, Ile to Val, Asn to Ser, Asp to Lys, and Ile to Thr at positions 76, 78, 117, 121, 125, 135, 137, 149, 162, 163, 173, 189, 195, and 199 respectively. The remaining 15 clinical isolates and the 6 raw sewage samples showed eight of the amino acid changes, at positions: 78, 121, 125, 135, 162, 173, 189, 195 and 199. All changes are similar to the amino acid changes of the three previously mentioned isolates (the two clinical isolates and an isolate from the raw sewage) except the amino acid change at position 195 showed a change of Asn to Asp.
Sequence analysis of human rotavirus VP8 gene from 18 Egyptian clinical isolates and 9 raw sewage samples which contained P[4] genotype (Tables S1 and S2) showed highest relation to the three human rotavirus A DS-1 reference strains (GenBank nucleotide accessions: EF672577.1, HQ650119.1 and DQ141310.1, GenBank protein accessions: ABV53252.1, AEG25325.1 and ABA18723.1 respectively) (Fig. 7) with 97.50–98.96% nucleotide identity and 96.88–100% amino acid identity (Table 3). Of the eighteen P[4] VP8 aligned sequenced clinical isolates, 15 isolates [collected during Nov. 2015 (2 isolates), Dec. 2015 (2 isolates), Jan. 2016 (4 isolates), Feb. 2016 (1 isolate), Nov. 2016 (2 isolates), Dec. 2016 (2 isolates) and Jan. 2017 (2 isolates), ID: EGY/SS18, SS19, SS20, SS22, SS24, SS25, SS26, SS27, SS28, SS29, SS30, S31, SS32, SS34, and SS35 respectively] and 8 raw sewage samples [Zenin Dec. 2015, Zenin Feb. 2016, El-Gabal El-Asfar Feb. 2016, El-Gabal El-Asfar Oct. 2016, El-Gabal El-Asfar Nov. 2016, El-Gabal El-Asfar Jan. 2017, Zenin January 2017, and Zenin Mar. 2017 (ID: EGY/ RSS8, RSS9, RSS10, RSS11, RSS12, RSS14, RSS15, and RSS16 respectively)] had similar sequences and showed five nucleotide substitutions in their coding sequences with 98.96% nucleotide identity and 100% homology in their amino acid sequences in comparison to the three human rotavirus A DS-1 reference strains (Table 3). The remaining three P[4] clinical isolates (collected during Dec. 2015, Jan. 2016, and Jan. 2017, ID: EGY S21, S23, and S33 respectively) and 1 raw sewage sample [El-Gabal El-Asfar Dec. 2016 (ID: EGY/RSS13)] had similar sequences and showed 12 nucleotide substitutions in their coding sequences, showed five amino acid changes (i.e. non-silent mutations: Lys to Asn, Lys to Asn, Asn to Glu, Asn to Lys, and Iel to Leu at positions: 75, 136, 150, 153, and 196 of the reference strains). They showed 97.50% nucleotide identity and 96.88% amino acid identity in comparison to the three human rotavirus A DS-1 reference strains (Table 3).
Table 3.
Comparison the similarities of nucleotide sequences and amino acids sequences between Egyptian rotavirus A VP8 P[4] partial gene (from Egyptian infants and raw sewage samples) and RVA reference strains P[4] sequences as well as the representative Rotarix® and Rotateq® strains
| Strain name | Genotype | Nucleotide accession | Protein accession | Nucleotide identity with the Egyptian sequences | Amino acids identity with the Egyptian sequences | ||
|---|---|---|---|---|---|---|---|
| EGY/SS18–SS20, SS22, SS24–SS32, SS34, SS35, RSS8–RSS12, RSS14–RSS16 (%) | EGY/SS21, SS23, SS33, RSS13 (%) | EGY/SS18–SS20, SS22, SS24-SS32, SS34, SS35, RSS8-RSS12, RSS14-RSS16 (%) | EGY/SS21, SS23, SS33, RSS13 (%) | ||||
| RVA/USA/DS-1/1976/G2P1B[4] | G2P[4] | EF672577.1 | ABV53252.1 | 98.96 | 97.50 | 100 | 96.88 |
| Rotavirus A RVA/DS-1/1976/G2P[4] | G2P[4] | HQ650119.1 | AEG25325.1 | 98.96 | 97.50 | 100 | 96.88 |
| Human rotavirus A isolate DS-1 | P[4] | DQ141310.1 | ABA18723.1 | 98.96 | 97.50 | 100 | 96.88 |
| RVA vaccine/USA/Rotarix® | G1P1A[8] | JN849113.1 | AEX30660.1 | 83.86 | 83.47 | 81.25 | 80.00 |
| RVA vaccine/USA/Rotateq® | G6P1A[8] | GU565044.1 | ADK26989.1 | 83.51 | 82.67 | 81.88 | 80.62 |
Alignment of the nucleotide sequences and amino acid sequences of the P[4] VP8 sequenced Egyptian clinical isolates and raw sewage samples with the vaccine strain Rotarix® showed 83.47–83.86% nucleotide identity and 80.00–81.25% amino acid identity. Alignment with the vaccine strain Rotateq® showed 82.67–83.51% nucleotide identity and 80.62–81.88% amino acid identity (Table 3).
Sequence analysis of human rotavirus VP8 gene from 10 Egyptian clinical isolates and 4 raw sewage samples which contained P[6] genotype (Tables S1 and S2) showed highest relation to the human rotavirus A reference strain 1076 (GenBank nucleotide accession: M88480.1, GenBank protein accession: AAA47337.1) (Fig. 7) with 98.12% nucleotide identity and 100% amino acid identity (Table 4). Although all the 10 P[6] VP8 aligned sequenced clinical isolates [collected during Oct. 2015 (1 isolate), Dec. 2015 (2 isolates), Jan. 2016 (3 isolates), Oct. 2016 (1 isolate), Dec. 2016 (1 isolate), Jan. 2017 (2 isolates), ID: EGY/SS36, SS37, SS38, SS39, SS40, SS41, SS42, SS43, SS44, and SS45 respectively] and 4 raw sewage samples [El-Gabal El-Asfar Oct. 2015, Zenin Nov. 2016, Zenin Dec. 2016, and El-Gabal El-Asfar Feb. 2017 (ID: EGY/RSS17, RSS18, RSS19, and RSS20 respectively)] which had similar sequences showed 9 nucleotide substitutions of the coding sequence, all mutations were silent.
Table 4.
Comparison the similarities of nucleotide sequences and amino acids sequences between Egyptian rotavirus A VP8 P[6] partial gene (from Egyptian infants and raw sewage samples) and RVA reference strain P[6] sequences as well as the representative Rotarix® and Rotateq® strains
| Strain name | Genotype | Nucleotide accession | Protein accession | Nucleotide identity with the Egyptian sequences | Amino acids identity with the Egyptian sequences |
|---|---|---|---|---|---|
| EGY/SS36-SS45, RSS17–RSS20 (%) | EGY/SS36-SS45, RSS17–RSS20 (%) | ||||
| Human rotavirus strain 1076 | P[6] | M88480.1 | AAA47337.1 | 98.12 | 100 |
| RVA vaccine/USA/Rotarix® | G1P1A[8] | JN849113.1 | AEX30660.1 | 67.58 | 62.50 |
| RVA vaccine/USA/Rotateq® | G6P1A[8] | GU565044.1 | ADK26989.1 | 68.83 | 62.50 |
Alignment of the nucleotide sequences and amino acid sequences of the P[6] VP8 sequenced Egyptian clinical isolates and raw sewage samples with the vaccine strain Rotarix® showed 67.58% nucleotide identity and 62.50% amino acid identity. Furthermore, alignment with the vaccine strain Rotateq® showed also non-significant similarity in both nucleotides (68.83%) and amino acids (62.50%) sequences (Table 4).
A neighbor joining phylogenetic tree of partial VP8 P[8] (477 bp), VP8 P[4] (480 bp) and VP8 P[6] (480 bp) gene sequences of group A rotaviruses from 17, 18, and 10 Egyptian clinical cases respectively and from 7, 9, and 4 Egyptian raw sewage samples respectively showed that P[8], P[4], and P[6] formed separate clusters as shown in Fig. 7.
Discussion
As universal rotavirus vaccination continues to increase worldwide, global surveillance of rotavirus has become an important focus of the vaccination programs. Rotavirus surveillance is vital to describe genotype distribution in different countries and their regions, and to discover potential emergence of new strains that possess neither VP7 nor VP4 specificity with the available rotavirus vaccines currently in use which may represent a challenge to the outcome and success of vaccination (Desai et al. 2012; Luches et al. 2015; Intamaso et al. 2017; Yu et al. 2019).
Several gastrointestinal cases from most Egyptian Governorates are frequently admitted to Abo El-Reech hospital, Greator Cairo, Egypt. Thus, this hospital can be used as an excellent model for rotavirus prevalence in Egyptian infants and children. In the present study, the frequency of rotavirus in diarrheal samples which collected from Abo El-Reech hospital from Oct. 2015 to Sep. 2017 was 24.37%. Rotaviruses were detected with higher frequency during autumn and winter months. A higher occurrence of group A rotavirus infections was observed from Nov. 2015 to Jan. 2016 and from Nov. 2016 to Jan. 2017 with highest percentage of prevalence rate of up to 35.90% and 35.59% in Jan. 2016 and Jan. 2017 respectively. The infection rate decreased during spring and summer months suggesting that the rotavirus group A infection rate was low and that gastroenteritis during this period may be caused by other pathogens. These results were confirmed with several previous studies (Dhital et al. 2017; Saudy et al. 2017). In another study, prevalence of rotavirus group A was investigated in 92 stool specimens collected from Mansoura University Children's Hospital, Egypt (Sep. 2010 to Feb. 2012) by RT-PCR. Rotavirus group A was detected in 48.90% of the cases. The Rotavirus negative cases (51.10%) included 34 (37.00%) patients had bacterial pathogens and 13 patients (14.10%) had no detectable pathogen. They found rotavirus diarrhea to be occurred mostly in the autumn and winter months, while most rotavirus negative cases were recorded in summer (Saudy et al. 2017). The present results agree also with those of Dhital et al. (2017) who collected 717 diarrheal stool samples from two hospitals situated in Kathmandu city, capital of Nepal, from January to November 2014. They detected rotavirus in 22.90% (161/717) of patients. The highest number of diarrhea was seen in January.
Detection of rotavirus with higher rates in males (26.27%) than in females (21.07%) was noted in the present study, however the statistical analysis using the Person Chi-square test indicated that no statistically significant differences could be established. This is in agreement with (Saluja et al. 2014) who suggested no gender preference for rotavirus infection, however other reports indicated that male children had been found to be significantly more susceptible (X2 = 2.97, P < 0.05) and likely to be admitted to hospitals than female children (Junaid et al. 2011).
Although the age of patients suffered from gastroenteritis in this study was up to 5 years, all of the infected children in the present study were ≤ 2 years of age with the highest prevalence of rotavirus (30.59%) in the age group of ]6, 12] months. This age distribution is in agreement with (Junaid et al. 2011) who reported that most of the infected children were under 2 years of age with the highest prevalence between 7–12 months. In the present study, rotavirus positivity rate was lower among children ≤ 6-month-old (18.31%). This lower incidence rate might be due to passive immunity acquired by the infants from their mothers which wades off after 6 months. This was also confirmed by (Rodrigues et al. 2007). In the present study, rotavirus infections were not observed in children older than 24 months. These may be attributed to the lower number of specimens collected from children older than 2 years. Another probability may be that infections with rotavirus decrease with increasing age over two years. Dian and co-workers (2017) noted that children less than 2 years of age were accounted for 87.40% (4712/5394) of the total infections and the infection rate showed an overall declining trend with increasing age after 36 months. Although rotavirus group A infections were also found on 5- to 14-year-old children, these accounted for only 2.00% (110/5394) of all positive samples.
Although several studies and for several years suggested that rotavirus group A infections were very frequent in children suffering from gastroenteritis with age up to 5 years, the recent studies including our present study limited and restricted the patterns of infections within these 5 years. This may help the physician and the workers in the hospitals to determine the cause of gastroenteritis of in-patient and out-patient children using rapid tests like agglutination and accurate tests like RT-PCR (El-Senousy et al. 2013c, 2015). Additionally, patient symptoms, patient age, and the time of infection during the year may help in the accurate diagnosis of the pathogens responsible for gastroenteritis (either bacteria, parasites, or viruses).
Our results of studying rotavirus G and P genotyping in positive rotavirus VP6 clinical specimens showed significant higher prevalence of common P genotypes than common G genotypes and this may reflect their prevalence in the Egyptian community. The significant higher prevalence of common P genotypes than common G genotypes was also observed in the raw sewage samples. The raw sewage samples were collected in autumn and winter months which represent the peak of prevalence of rotavirus in the aquatic environment either in Egypt (Villena et al. 2003; El-Senousy et al. 2004, 2013a, 2014a, b, 2015; El-Senousy and El-Mahdy 2009) or worldwide (Li et al. 2011; Barril et al. 2015; Zhou et al. 2016). It may give an indication about the significant higher prevalence of uncommon G genotypes than uncommon P genotypes in Egyptian clinical and environmental samples and consequently in the Egyptian community. The higher prevalence of common P genotypes than common G genotypes in Egyptian infants agree with reports from other African countries but with different proportion in prevalence (Ouermi et al. 2017; Lartey et al. 2018; Tagbo et al. 2019). Ouermi et al. (2017) provided a comprehensive view of the current circulating rotavirus strains in Africa (2006–2016) which is important in light of the new rotavirus vaccinations. They reported that almost all the African regions showed the same predominant genotypes with some variations in prevalence. Genotypes G1, G2, G3, G9 and G12 accounted for 75.94% of all G genotypes circulating in Africa, whereas genotypes P[4], P[6], and P[8] were predominant and accounted for 82.94% of the P genotypes encountered. The difference in circulating strains is not surprising given that genotypes vary much from one country to another and from one region to another. In Ghana, Lartey and co-workers (2018) studied the molecular characterization of 1363 rotavirus positive diarrheic stool samples (from children under-five-years old) from Jan. 2009 to Dec. 2016. The most common G genotypes detected during the entire period were G1 (30.70%), G12 (13.80%), G3 (12.10%) and G2 (10.70%), while the most commonly detected P genotypes were P[8] (37.80%), P[6] (31.60%), and P[4] (8.90%). In Mozambique, 157 rotavirus positive clinical specimens collected between 2012 and 2013 from children under-five-years old were tested for G and P genotyping prior to vaccine introduction. Genotypes G2 (32.40%), G12 (28.00%), P[4] (41.40%), and P[6] (22.90%) strains were commonly detected (Joao et al. 2018). The higher prevalence of common P genotypes than common G genotypes in Egyptian raw sewage samples agrees with the results reported by (El-Senousy and Abou-Elela 2017) who studied the rotavirus genotypes in a pilot wastewater treatment plant in Egypt and their results showed that G1 was the most frequent common G genotype while P[8] and P[4] were the most frequent common P genotypes in the sewage samples. It agrees also with reports from Tunisia where rotavirus group A could be genotyped for G and P genotypes in 228 and 247 out of 375 rotavirus positive sewage samples respectively. Genotypes G1 (29.10%), P[8] (32.00%), and P[4] (27.70%) were commonly detected (Hassine-Zaafrane et al. 2015). In Argentina, rotavirus G genotypes and P genotypes could be characterized in 29 (82.86%) and 22 (62.80%) respectively out of the 35 raw sewage samples collected monthly from Feb. 2009 to Dec. 2011. The most common G genotypes detected during the entire period were G1 (32.20%), G2 (23.70%), and G9 (22.00%), while the most common P genotypes detected were P[8] (67.90%) and P[4] (32.10%) (Barril et al. 2015). Our results showed a strong relationship between the prevalence of common P and G genotypes in both clinical specimens and raw sewage samples. Common P genotypes were significantly higher than common G genotypes in the positive rotavirus VP6 clinical specimens and raw sewage samples. The percentage of common P genotypes in the raw sewage samples (87.50%) was increased in comparison to the results of Villena and co-workers (2003) which common P genotypes were detected in 73.33% of the positive raw sewage samples collected from Greater Cairo in 1998–1999. P[4] genotype was the most prevalent genotype in our results followed by P[8] and finally P[6] in both clinical specimens and environmental samples although it was next to P[8] and P[6] genotypes in the results of Villena and co-workers (2003). Increasing the percentage of frequency of P[4] genotype was observed in the Egyptian investigated samples (clinical and environmental) through 20 years (1998–2017) in several studies (Villena et al. 2003; El-Senousy et al. 2004, 2014a; El-Esnawy et al. 2010; El-Senousy and Abou-Elela 2017). This may return to the ability for reassortment of the rotavirus genome or increasing the rate of infection with rotavirus strains contained genotype P[4] in the Egyptian community. The rate of viral presence in the aquatic environment as a result of its presence in the community (infected or carrier persons) and also its resistance to environmental conditions and consequently its long persistence may increase the chance for genome reassortment. All these conditions were observed through a lot of studies concerned with rotaviruses in the Egyptian aquatic environment (Villena et al. 2003; El-Senousy et al. 2004, 2013a, 2014a, b, c, d, 2015; El-Senousy and El-Mahdy 2009; El-Senousy and Abou-Elela 2017). The increased frequency of P[4] genotype in the Egyptian clinical and environmental samples may be the reason of increasing the percentage of the common P genotypes in the clinical and environmental samples in our study. The percentage of common G genotypes in the raw sewage samples (54.17%) was decreased in comparison to the results of Villena and co-workers (2003) which common G genotypes were detected in 63.91% of the positive rotavirus VP6 raw sewage samples. This may return to increasing the percentage of uncommon G genotypes which G9 was detected in our results in 16.67% of the rotavirus VP6 positive raw sewage samples, although it was detected in 12.43% of the rotavirus VP6 positive samples in the study of Villena and co-workers (2003) which G9 was detected as an emergence of a new strain either in Egyptian clinical or environmental samples. This was supported by our results of clinical specimens which G9 was detected in 31 specimens (12.40%). Decreasing the rate of infection with the common G genotypes in the Egyptian community may be another reason. Radwan and co-workers (1997) reported that 39.00% of the clinical specimens collected from Cairo (1992–1993) were untypeable for common G genotypes. Although it is a high percentage of untypeable G specimens, it is less than the percentage of untypeable G specimens in our present study. G1 was the highest prevalent common G genotype through all the previous studies concerned with rotaviruses in the aquatic environment and in our present study in both clinical specimens and raw sewage samples. G3 was second to G1 in the present study in both clinical specimens and raw sewage samples. In the study of Villena and co-workers (2003), the prevalence of G3 was also second to G1 in the raw sewage samples of Greater Cairo in 1998–1999. Raw sewage represents the reservoir of the enteric viruses (pathogens). It receives the feces contained enteric viruses (pathogens) excreted by infected persons or carriers. Raw sewage also could be considered as a source of infection for other persons by contamination of either drinking water resources or irrigation water and consequently freshly eaten crops (El-Senousy et al. 2013b, 2013e). Raw sewage could be considered as a mirror reflects the levels and types of viruses (pathogens) circulating in the community.
In 2006, pivotal clinical trials of 2 live oral rotavirus vaccines—a pentavalent bovine–human reassortant vaccine given in a 3-dose schedule (Rotateq®, Merck & Co), and a monovalent human vaccine given in a 2-dose schedule (Rotarix®, GSK Biologicals)- demonstrated good efficacy (85.00–98.00%) in preventing severe rotavirus gastroenteritis in developed countries (Ruiz-Palacios et al. 2006; Vesikari et al. 2006). Subsequent trials in Africa showed a more modest efficacy (59.00–64.00%) but given the large disease burden in these regions, the vaccine prevented more disease than in lower burden settings (Armah et al. 2010; Madhi et al. 2010). Reports from different African countries showed that the RotaTeq® vaccine efficacy for the complete follow up differed by country. Kenya had the highest point estimate of efficacy, followed by Ghana, and then Mali. Overall efficacy against severe rotavirus gastroenteritis was more than 64% in the first year of life, and more than 19% in the second year of life. As for the analysis of primary outcomes, vaccine efficacy in the first year of life was similar between Kenya and Ghana, but was lower in Mali than in the other two countries. Overall, 148 of 189 infants in the vaccine group had IgA seroresponses (a more than three-fold rise from before dose one to after dose three) after completion of the vaccine series. The IgA seroresponse in the pentavalent rotavirus vaccine group was 78.90% in Ghana, 73.80% in Kenya, and 82.50% in Mali. Geometric mean titres after the third dose were similar in the vaccine groups from every country. Geometric mean titres of serum neutralizing antibodies against human rotavirus serotypes in the vaccine were 33.10 for serotype G1, 29.10 for serotype G2, 15.00 for serotype G3, 49.70 for serotype G4, and 44.90 for serotype P[8]. However, seroresponse rates for serum neutralizing antibodies (i.e. a three-fold or higher rise from baseline to after dose 3) were low in this trial, in which infants had high concentrations of serum neutralizing antibodies before the first dose of vaccine or placebo for the rotavirus serotypes contained in the vaccine (Armah et al. 2010). In the study performed on Malawi's infants, Rotarix® vaccine effectiveness for two doses was 64.00% in test-negative control individuals and 63.00% in community controls. For children with more severe disease, effectiveness for two doses of Rotarix® vaccine was 68.00% in test-negative control individuals and 68.00% in community controls. The two-dose vaccine effectiveness point estimate was higher for rotavirus G1 (82.00% and 78.00% for test negative and community controls respectively), than for rotavirus G2 (53.00% and 61.00% respectively), or for G12 (53.00% and 61.00% respectively) (Bar-Zeev et al. 2015). Bar-zeev and co-workers (2015) showed robust effectiveness of the G1P[8] Rotarix® vaccine despite genotypic rotavirus diversity in the population, with fully homotypic (G1P[8]), fully heterotypic (G2P[4], G2P[6], and G12P[6]), and partially heterotypic (G12P[8]) genotypes circulating during their study. The problems which may be associated with the live attenuated vaccines such as the risk of reversion and intussuscptions and other factors such as the high cost of the vaccine may reduce vaccine use (Desai et al. 2012; Madsen et al. 2012). These reasons could also limit their applications for Egyptian infants. Thus non-living recombinant subunit vaccine is therefore considered as an alternative for rotavirus immunization. Recombinant subunit vaccines are made from a fragment of protein expressed in the laboratory using the viral gene. A lot of reports showed the advantages of the recombinant subunit vaccines in relation to the live attenuated vaccines, such as safety profile (due to the lack of replication) and production cost, although they may require the use of adjuvants to elicit a protective and long-lasting immune response (Nascimento and Leite 2012; Perez et al. 2012; El-Senousy et al. 2013d; Chen et al. 2017). The two licensed live attenuated oral rotavirus vaccines do not cover all human rotavirus common P genotypes (such as P[4] and P[6]) circulated in the Egyptian rotavirus cases and in the raw sewage samples in the present study. The currently licensed RotaTeq® vaccine is based on the most common G genotypes (G1-4)P[5] and G6P[8]) while Rotarix® is based on G1P[8]. The surveillance of common G and P genotypes in clinical specimens and environmental samples contributes to a better understanding of rotavirus in circulation and helps to characterize the various antigenic shifts that could reduce vaccine efficacy.
It had been well established that rotavirus G-P combinations G1P[8], G2P[4], G3P[8], and G4P[8] are of global epidemiologic importance with G1P[8] being the most important (Santos and Hoshino 2005). Currently available two live oral rotavirus vaccines were selected or designed to provide coverage to such epidemiologically important G and P genotypes since data obtained from experimental animal studies as well as vaccine clinical trials and volunteer studies appeared to suggest that the induction of serotype-specific immunity may be important for optimal protection (Kapikian and Hoshino 2007; Linhares et al. 2008). It was of note that unusual G and P genotypes causing a high incidence of human infection had been reported in various regions of the world including G8, G9, G10, and G12 (Holmes et al. 1999; Steele and Ivanoff 2003; Rahman et al. 2007; Matthijnssens et al. 2008; Sanz et al. 2009; Castello et al. 2009) that are not covered serotypically by Rotarix® and RotaTeq®. In our study, high percentage of untypeable samples for common G genotypes may indicate high percentage of uncommon G genotypes. Since in nature, almost all human rotavirus G genotypes had been detected in combination with P[8], P[4], or P[6] (Wen et al. 2012). Based on that Wen and co-workers (2012) suggested that P[8], P[4], and P[6] ∆VP8* proteins which generated in their study could be used at a minimum singly or preferably in multivalent formulations of two or more components to provide antigenic coverage to almost all the G (VP7) types of global as well as regional epidemiologic importance. In our study, amplification of VP8 fragments of P[8], P[4], and P[6] genotypes which encode amino acid residues 65–223 of Wa VP8, 64–223 of DS-1 VP8, and 64–223 of 1076 VP8 was performed. Wen and co-workers (2012) reported that guinea pigs hyperimmunized with purified Wa P[8] ∆VP8* elicited high levels of VP8*-homotypic (range 1:7680–1:5120) as well as VP8*-heterotypic (1:5120 vs P[4]) neutralizing antibodies. In addition, low levels (1:320 and 1:80) of VP8*-heterotypic neutralizing antibodies were induced against P[6] and P[10] respectively. Similarly, guinea pigs hyperimmunized with purified DS-1 P[4] ∆VP8* elicited high levels of homotypic (1:5120) and heterotypic (1:1280 vs P[8]), as well as moderate to low levels (1:320 and 1:80 vs P[6] and P[10] respectively) neutralizing antibodies. Guinea pigs hyperimmunized with 1076 P[6] ∆VP8*, developed high levels of VP8*-homotypic (1:1280) as well as lower heterotypic neutralizing antibodies (1:320 to 1:40 vs P[8] and P[10] respectively). It was of interest that these serological relationships among four different P type strains (i.e. P[4], P[6], P[8], and P[10]) correlated with molecular relationships among them: Wa P[8] strain shares the highest amino acid identity of 82.40% with DS-1 P[4] strain followed by 64.20% with 1076 P[6] strain and 46.80% with 69 M P[10] strain. Mice immunized with DS-1 ∆VP8* developed high levels of homotypic (i.e. DS-1 P[4] ∆VP8*) as well as moderate to low levels of heterotypic (i.e. Wa P[8] ∆VP8*) IgG antibodies. In addition, selected mouse sera showed both homologous (1:80 vs Wa strain) neutralizing activities. These data indicated that these selected ΔVP8 proteins may be a possible additional candidate as new parenteral rotavirus vaccines. In case of Egyptian clinical specimens, our results showed that 89.20% of the clinical specimens contained at least one of the common P genotypes (P[8], P[4], and P[6]). These P genotypes were combined with typeable common G genotypes (G1 or G3) or untypeable G genotypes. The percentage of untypeable P cases but typeable for G3 was 3.20%, while the percentage of untypeable P cases but typeable for G1 was 5.60%. This may increase the efficiency of the recombinant subunit vaccine which includes the specific proteins of the common P genotypes if it includes also the protein of either G1 or G3 genotypes. These results were supported also by the results of the prevalence of common P genotypes in the raw sewage samples. Common P genotypes were detected in 83.33% of the rotavirus VP6 positive raw sewage samples. Additionally, one raw sewage sample was untypeable for common P genotypes but contained G1 genotype (4.17%). The significantly lower value of the percentage of the common G-typeable clinical specimens in addition to the percentage of P[8] or P[4] or P[6] in the common G-untypeable clinical specimens than the percentage of the common P-typeable clinical specimens in addition to the percentage of G1 or G3 in the common P-untypeable clinical specimens indicated an advantage to the higher prevalence combinations as targets for recombinant subunit vaccine. This was also supported by the results of the raw sewage samples.
In the present study, sequence analysis of human rotavirus VP8 gene of the 17 clinical isolates and the 7 raw sewage isolates which contained P[8] genotype showed 97.06–99.16% nucleotide identity and 95.60–100% amino acid identity with the two human rotavirus reference strains (Wa strain and rotavirus A strain USA). Of the seventeen P[8] VP8 sequenced clinical isolates and of the seven P[8] VP8 sequenced raw sewage samples, two clinical isolates and one raw sewage isolate showed seven non-silent mutations in the amino acid sequences in comparison to the two reference strains while the remaining clinical isolates and raw sewage isolates showed silent mutations. Furthermore, sequence analysis of human rotavirus VP8 gene of the 18 clinical isolates and the 9 raw sewage isolates which contained P[4] genotype showed 97.50–98.96% nucleotide identity and 96.88–100% amino acid identity with the three human rotavirus DS-1 reference strains. Of the eighteen P[4] VP8 sequenced clinical isolates and the nine P[4] VP8 sequenced raw sewage isolates, three clinical isolates and one raw sewage isolate showed five non-silent mutations at five amino acid positions in comparison to the three reference strains while the remaining clinical isolates and raw sewage isolates showed silent mutations. Moreover, sequence analysis of human rotavirus VP8 gene of the 10 clinical isolates and the 4 raw sewage isolates which contained P[6] genotype showed 98.12% nucleotide identity and 100% amino acid identity with the human rotavirus reference strain (strain 1076). Although the ten clinical isolates and the four raw sewage isolates showed 9 nucleotide substitutions in the coding sequence, all mutations were silent. The low variations in the amino acids of the Egyptian clinical and environmental VP8 sequences and the lower number of samples contained non-silent mutations [5/45 (11.11%)] in the sequences of the clinical specimens and [2/20 (10%)] in the sequences of the raw sewage samples in comparison to the reference strains in the VP8 partial gene may put the ∆VP8 proteins of the ∆VP8 partial gene (nt 202 to nt 678 for Wa P[8], nt 190 to nt 669 for DS-1 P[4], and nt 199 to nt 678 for 1076 P[6]) as a possible additional candidate as future recombinant subunit rotavirus vaccine. Addition of the protein of either G1 or G3 may increase the vaccine efficiency.
The P[8] VP8 sequenced clinical and environmental isolates showed 93.71–98.11% amino acid identity with that of the Rotarix® vaccine strain while they showed lower amino acid identity (91.19–94.34%) with Rotateq® vaccine strain. Of the seventeen P[8] VP8 sequenced isolates, two isolates collected during Jan. 2016 and Nov. 2016 in addition to one raw sewage sample (El-Gabal El-Asfar, Jan.2016) were identical to each other and showed 10 non-silent mutations in the amino acid sequences in comparison to that of the Rotarix® vaccine strain while the remaining 15 clinical isolates and 6 raw sewage isolates showed 3 non-silent mutations. Likewise, the two clinical isolates collected during Jan. 2016 and Nov. 2016 and the raw sewage isolate collected during Jan. 2016 from El-Gabal El-Asfar WWTP showed 14 non-silent mutations in the amino acid sequences in comparison to that of the RotaTeq® vaccine strain while the remaining 15 clinical isolates and 6 raw sewage isolates showed 8 non-silent mutations. It had been known that Rotarix® VP8 belongs to P[8] lineage 1 while RotaTeq® VP8 belongs to lineage 2 (Kulkarni et al. 2014). This may explain the lower similarity in the amino acid sequences of VP8 P[8] Egyptian clinical and raw sewage isolates of the present study in comparison to the RotaTeq® vaccine and the higher similarity in comparison to the Rotarix® vaccine.
Phylogenetic analysis of the present study confirmed that P[8], P[4], and P[6] form separate clusters and this may interpret the lower similarity of P[4] VP8 amino acid sequences in comparison to the VP8 P[8] vaccine strains (80.00–81.25% with Rotarix® and 80.62–81.88% with RotaTeq®) and the non-significant similarity of P[6] VP8 (62.50%) amino acid sequences in comparison to the VP8 P[8] vaccine strains. This finding was also confirmed by Zeller et al. (2012) who indicated that the antigenic epitopes of rotavirus A strains contained in the vaccines were significantly different from those of the rotavirus group A strains circulating in Belgium.
Future studies will be needed to develop a safe non-living recombinant subunit vaccine based on common rotavirus G and P genotypes circulating in the Egyptian community. Studying the effect of non-silent mutations of different P genotypes on the efficiency of the developed vaccines must be included. Also, investigation of silent and non-silent mutations in different common G genotypes must be performed. Future research will be needed also to study the effect of silent and non-silent mutations in different P and G genotypes on the efficiency of the present vaccines (Rotarix® and RotaTeq®).
As general conclusions, the study showed high burden of rotavirus gastroenteritis as well as higher prevalence of common P genotypes than common G genotypes in Egyptian infants and raw sewage samples. Thus, the prevalence of rotavirus genotypes that are not covered by the currently available live attenuated oral rotavirus vaccines will probably reduce the efficacy of these rotavirus vaccines on Egyptian infants. High prevalence of common P genotypes in any community may put the ΔVP8-specific proteins as a possible additional candidate as future recombinant subunit rotavirus vaccine especially in some communities which have significantly higher prevalence of common P genotypes than common G genotypes. Addition of the protein of the most prevalent G genotype in the community to the vaccine may increase its efficiency. The most common rotavirus genotypes circulating in the different communities may be taken into consideration in the future vaccine development. The low percentage of the non-silent mutations in the human rotavirus VP8 partial gene may give an additional advantage to the future developed recombinant subunit vaccine.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge In-House Project Office, National Research Centre (NRC), Egypt, for financial support of this work, Project No.: 11090332, PI: Prof. Dr. Waled Morsy El-Senousy.
Compliance with Ethical Standards
Ethical Approval
Ethical approval was obtained from the head of gastroenteritis unit of Abo El-Reech hospital.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- APHA (American Public Health Association) Standard methods for the examination of water and wastewater. 23. Washington, DC: American Public Health Association; 2017. [Google Scholar]
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub–Saharan Africa: A randomised, double–blind, placebo–controlled trial. Lancet. 2010;376(9741):606–614. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, et al. Family Reoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, et al., editors. Virus taxonomy: Classification and nomenclature: Ninth report of the International Committee on Taxonomy of Viruses. San Diego, California: Elsevier Academic Press; 2012. pp. 541–637. [Google Scholar]
- Bányai K, László B, Duque J, Steele AD, Nelson EA, Gentsch JR, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: Insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(1):A122–A130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- Barril PA, Fumian TM, Prez VE, Gil PI, Martínez LC, Giordano MO, et al. Rotavirus seasonality in urban sewage from Argentina: effect of meteorological variables on the viral load and the genetic diversity. Environmental Research. 2015;138:409–415. doi: 10.1016/j.envres.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Bar-Zeev N, Kapanda L, Tate JE, Jere KC, Iturriza-Gomara M, Nakagomi O, et al. Effectiveness of a monovalent rotavirus vaccine in infants in Malawi after programmatic rol-out: An observational and case–control study. The Lancet Infectious Diseases. 2015;15(4):422–428. doi: 10.1016/S1473-3099(14)71060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin JB, Macartney KK, Lee KJ, Quinn HE, Buttery J, Lopert R, et al. Intussusception risk and disease prevention associated with rotavirus vaccines in Australia’s national immunization program. Clinical Infectious Diseases. 2013;57(10):1427–1434. doi: 10.1093/cid/cit520. [DOI] [PubMed] [Google Scholar]
- Castello AA, Nakagomi T, Nakagomi O, Jiang B, Kang JO, Glass RI, et al. Characterization of genotype P[9]G12 rotavirus strains from Argentina: High similarity with Japanese and Korean G12 strains. Journal of Medical Virology. 2009;81(2):371–381. doi: 10.1002/jmv.21384. [DOI] [PubMed] [Google Scholar]
- Chen SC, Tan LB, Huang LM, Chen KT. Rotavirus infection and the current status of rotavirus vaccines. Journal of the Formosan Medical Association. 2012;111:183–193. doi: 10.1016/j.jfma.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Chen YC, Cheng HF, Yang YC, Yeh MK. Biotechnologies applied in biomedical vaccines. In: Afrin F, Hemeg H, Ozbak H, editors. Vaccine. Rijeka: Intech Open; 2017. pp. 97–110. [Google Scholar]
- Desai R, Curns AT, Patel MM, Parashar UD. Trends in intussusceptions associated deaths among US infants from 1979–2007. The Journal of Pediatrics. 2012;160(3):456–460. doi: 10.1016/j.jpeds.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Dhital S, Sherchand JB, Pokhrel BM, Parajuli K, Shah N, Mishra SK, et al. Molecular epidemiology of rotavirus causing diarrhea among children less than five years of age visiting national level children hospitals, Nepal. BMC Pediatrics. 2017;17(1):101–107. doi: 10.1186/s12887-017-0858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian Z, Fan M, Wang B, Feng Y, Ji H, Dong S, et al. The prevalence and genotype distribution of rotavirus A infection among children with acute gastroenteritis in Kunming, China. Archives of Virology. 2017;162:281–285. doi: 10.1007/s00705-016-3102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz R, Kalaycioglu AT, Acar S, Bakkaloglu Z, Karagoz A, Korukluoglu G, et al. Prevalence of rotavirus genotypes in children younger than 5 years of age before the introduction of a universal rotavirus vaccination program: Report of rotavirus surveillance in Turkey. PLoS ONE. 2014;9(12):e113674. doi: 10.1371/journal.pone.0113674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Esnawy NA, El-Senousy WM, Hammad IA, Abada EA, Abu-Zekry M, Rizk NM. Epidemiology of rotavirus in Greater Cairo. The New Egyptian Journal of Medicine. 2010;42(1):43–51. [Google Scholar]
- El-Senousy WM, Abou-Elela SI. Assessment and evaluation of an integrated hybrid anaerobic–aerobic sewage treatment system for the removal of enteric viruses. Food and Environmental Virology. 2017;9(3):287–303. doi: 10.1007/s12560-017-9286-4. [DOI] [PubMed] [Google Scholar]
- El-Senousy WM, Barakat AB, Ghanem HE, Kamel MA. Molecular epidemiology of human adenoviruses and rotaviruses as candidate viral indicators in the Egyptian sewage and water samples. World Applied Sciences Journal. 2013;27(10):1235–1247. [Google Scholar]
- El-Senousy WM, Costafreda MI, Pintó RM, Bosch A. Method validation for norovirus detection in naturally contaminated irrigation water and fresh produce. International Journal of Food Microbiology. 2013;167:74–79. doi: 10.1016/j.ijfoodmicro.2013.06.023. [DOI] [PubMed] [Google Scholar]
- El-Senousy WM, El-Gamal MS, Kamel MM, El-Mahdy EM. Prevalence of human and animal rotaviruses and HEV in Egyptian Nile water resources. World Applied Sciences Journal. 2014;32(11):2218–2228. [Google Scholar]
- El-Senousy WM, El-Gamal MS, Mousa AAE, El-Hawary SE, Fathi MN. Prevalence of noroviruses among detected enteric viruses in Egyptian aquatic environment. World Applied Sciences Journal. 2014;32(11):2186–2205. [Google Scholar]
- El-Senousy WM, El-Gamal MS, Mousa AAE, El-Hawary SE, Kamel MM, Fathi MN, et al. Effect of chlorine on noroviruses, rotaviruses, and Hepatitis E virus in drinking water. World Applied Sciences Journal. 2014;32(11):2206–2212. [Google Scholar]
- El-Senousy WM, El-Mahdy EM. Detection and genotyping of rotaviruses in water treatment plants of El-Dakahlia Governorate. Egyptian Journal of Biotechnology. 2009;31:25–34. [Google Scholar]
- El-Senousy WM, Osman GA, Melegy AA. Survival of adenovirus, rotavirus, hepatitis A virus, pathogenic bacteria and bacterial indicators in ground water. World Applied Sciences Journal. 2014;29(3):337–348. [Google Scholar]
- El-Senousy, W. M., Pintó, R. M., & Bosch, A. (2004). Epidemiology of human enteric viruses in the Cairo water environment. Paper presented at the 1st International Conference of Environmental Research Division on Sustainable Development Environmental Challenges Facing Egypt. National Research Centre, Cairo, Egypt.
- El-Senousy WM, Ragab AME, Handak EMA. Rotaviruses group A and C in clinical samples. The New Egyptian Journal of Medicine. 2013;49(1):1–6. [Google Scholar]
- El-Senousy WM, Ragab AME, Handak EMA. Prevalence of rotaviruses groups A and C in Egyptian children and aquatic environment. Food and Environmental Virology. 2015;7(2):132–141. doi: 10.1007/s12560-015-9184-6. [DOI] [PubMed] [Google Scholar]
- El-Senousy WM, Shahein YE, Barakat AB, Ghanem HE, El-Hakim AE, Ameen SM. Molecular cloning and immunogenicity evaluation of rotavirus structural proteins as candidate vaccine. International Journal of Biological Macromolecules. 2013;59:67–71. doi: 10.1016/j.ijbiomac.2013.04.003. [DOI] [PubMed] [Google Scholar]
- El-Senousy WM, Sidkey NM, Abu Senna ASM, Abed NN, Hasan SF. Prevalence of rotaviruses and noroviruses in ground water of some rural areas in El-Giza Governorate. The New Egyptian Journal of Medicine. 2013;48(1):18–25. [Google Scholar]
- Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Iturriza-Gomara M, et al. Environmental monitoring for gastroenteric viruses in a pediatric primary immunodeficiency unit. Journal of Clinical Microbiology. 2006;44:395–399. doi: 10.1128/JCM.44.2.395-399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Glass RI, Woods PV, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. Journal of Clinical Microbiology. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. Journal of Clinical Microbiology. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V, Santos N, Timenetsky Mdo C. Identification of bovine and porcine rotavirus G types by PCR. Journal of Clinical Microbiology. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009;136:1939–1951. doi: 10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. The Journal of Infectious Diseases. 2017;215:34–41. doi: 10.1093/infdis/jiw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassine-Zaafrane M, Kaplon J, Ben Salem I, Sdiri-Loulizi K, Sakly N, Pothier P, et al. Detection and genotyping of group A rotaviruses isolated from sewage samples in Monastir, Tunisia between April 2007 and April 2010. Journal of Applied Microbiology. 2015;119:1443–1453. doi: 10.1111/jam.12920. [DOI] [PubMed] [Google Scholar]
- Holmes JL, Kirkwood CD, Gerna G, Clemens JD, Rao MR, Naficy AB, et al. Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Archives of Virology. 1999;144(7):1381–1396. doi: 10.1007/s007050050594. [DOI] [PubMed] [Google Scholar]
- Intamaso U, Poomipak W, Chutoam P, Chotchuang P, Sunkkham W, Srisopha S, et al. Genotype distribution and phylogenetic analysis of rotaviruses in Thailand and emergence of uncommon genotypes. Archives of Clinical Microbiology. 2017;8(4):60–70. [Google Scholar]
- Iturriza-Gómara M, Wong C, Blome S, Desselberger U, Gray J. Molecular characterization of VP6 genes of human rotavirus isolates: Correlation of genogroups with subgroups and evidence of independent segregation. Journal of Virology. 2002;76:6596–6601. doi: 10.1128/JVI.76.13.6596-6601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- João ED, Strydom A, O'Neill HG, Cuamba A, Cassocera M, Acácio S, et al. Rotavirus A strains obtained from children with acute gastroenteritis in Mozambique, 2012–2013: G and P genotypes and phylogenetic analysis of VP7 and partial VP4 genes. Archives of Virology. 2018;163(1):153–165. doi: 10.1007/s00705-017-3575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junaid SA, Umeh C, Olabode AO, Banda JM. Incidence of rotavirus infection in children with gastroenteritis attending Jos university teaching hospital, Nigeria. Virology Journal. 2011;8:233–240. doi: 10.1186/1743-422X-8-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian AZ, Hoshino Y. To serotype or not to serotype: that is still the question. The Journal of Infectious Diseases. 2007;195(5):611–614. doi: 10.1086/510862. [DOI] [PubMed] [Google Scholar]
- Karami M, Berangi Z. The need for rotavirus vaccine introduction in the national immunization program of more than 100 countries around the world. Infection Control and Hospital Epidemiology. 2018;39(1):124–125. doi: 10.1017/ice.2017.237. [DOI] [PubMed] [Google Scholar]
- Kulkarni R, Arora R, Arora R, Chitambar SD. Sequence analysis of VP7 and VP4 genes of G1P[8] rotaviruses circulating among diarrhoeic children in Pune, India: A comparison with Rotarix and RotaTeq vaccine strains. Vaccine. 2014;32(1):A75–A83. doi: 10.1016/j.vaccine.2014.03.080. [DOI] [PubMed] [Google Scholar]
- Lappalainen S, Pastor AR, Malm M, López-Guerrero V, Esquivel-Guadarrama F, Palomares LA, et al. Protection against live rotavirus challenge in mice induced by parenteral and mucosal delivery of VP6 subunit rotavirus vaccine. Archives of Virology. 2015;160(8):2075–2078. doi: 10.1007/s00705-015-2461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartey BL, Damanka S, Dennis FE, Enweronu-Laryea CC, Addo-Yobo E, Ansong D, et al. Rotavirus strain distribution in Ghana pre- and post-rotavirus vaccine introduction. Vaccine. 2018;36(47):7238–7242. doi: 10.1016/j.vaccine.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Gu AZ, Zeng SY, Yang W, He M, Shi HC. Monitoring and evaluation of infectious rotaviruses in various wastewater effluents and receiving waters revealed correlation and seasonal pattern of occurrences. Journal of Applied Microbiology. 2011;110(5):1129–1137. doi: 10.1111/j.1365-2672.2011.04954.x. [DOI] [PubMed] [Google Scholar]
- Li K, Lin XD, Huang KY, Huang KY, Zhang B, Shi M, et al. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology. 2016;494:168–177. doi: 10.1016/j.virol.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhares AC, Velazquez FR, Perez-Schael I, Saez-Llorens X, Abate H, Espinoza F, et al. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: A randomised, double-blind, placebo-controlled phase III study. Lancet. 2008;371(9619):1181–1189. doi: 10.1016/S0140-6736(08)60524-3. [DOI] [PubMed] [Google Scholar]
- Luchs A, Cilli A, Morillo SG, Carmona RC, Timenetsky MC. Rotavirus genotypes circulating in Brazil, 2007–2012: Implications for the vaccine program. Revista do Instituto de Medicina Tropical de São Paulo. 2015;57(4):305–313. doi: 10.1590/S0036-46652015000400006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. The New England Journal of Medicine. 2010;362:289–298. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- Madsen LB, Ustrup M, Fischer TK, Bygbjerg IC, Konradsen F. Reduced price on rotavirus vaccines: Enough to facilitate access where most needed? Bulletin of the World Health Organization. 2012;90(7):554–556. doi: 10.2471/BLT.11.094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Mcdonald SM, Attoui H, Bányai K, Brister JR, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Archives of Virology. 2011;156(8):1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Rahman M, Van Ranst M. Two out of the 11 genes of an unusual human G6P[6] rotavirus isolate are of bovine origin. Journal of General Virology. 2008;89(10):2630–2635. doi: 10.1099/vir.0.2008/003780-0. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Van Ranst M. Genotype constellation and evolution of group A rotaviruses infecting humans. Current Opinion in Virology. 2012;2(4):426–433. doi: 10.1016/j.coviro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Nascimento IP, Leite LC. Recombinant vaccines and the development of new vaccine strategies. Brazilian Journal of Medical and Biological Research. 2012;45:1102–1111. doi: 10.1590/S0100-879X2012007500142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouermi D, Soubeiga D, Nadembega WMC, Sawadogo PM, Zohoncon TM, Obiri-Yeboah D, et al. Molecular epidemiology of rotavirus in children under five in Africa (2006–2016): a systematic review. Pakistan Journal of Biological Science. 2017;20(2):59–69. doi: 10.3923/pjbs.2017.59.69. [DOI] [PubMed] [Google Scholar]
- Parashar UD, Johnson H, Steele AD, Tate JE. Health impact of rotavirus vaccination in developing countries: Progress and way forward. Clinical Infectious Diseases. 2016;62(2):S91–S95. doi: 10.1093/cid/civ1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez O, Batista-Duharte A, Gonzalez E, Zayas C, Balboa J, Cuello M, et al. Human prophylactic vaccine adjuvants and their determinant role in new vaccine formulations. Brazilian Journal of Medical and Biological Research. 2012;45:681–692. doi: 10.1590/S0100-879X2012007500067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwane SF, Gabr MK, El-Maraghi S, El-Saifi AF. Serotyping of group A rotaviruses in Egyptian neonates and infants less than 1 year old with acute diarrhea. Journal of Clinical Microbiology. 1997;35:2996–2998. doi: 10.1128/jcm.35.11.2996-2998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Sultana R, Ahmed G, Nahar S, Hassan ZM, Saiada F, et al. Prevalence of G2P[4] and G12P[6] rotavirus, Bangladesh. Emerging Infectious Diseases. 2007;13(1):18–24. doi: 10.3201/eid1301.060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A, de Carvalho M, Aaby P. Hospital surveillance of rotavirus infection and nosocomial transmission among children in Guinea-Bissau. The Pediatric Infectious Disease Journal. 2007;26(3):233–237. doi: 10.1097/01.inf.0000254389.65667.8b. [DOI] [PubMed] [Google Scholar]
- Rui-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, Clemens SC, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. The New England Journal of Medicine. 2006;354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- Saluja T, Sharma SD, Gupta M, Kundu R, Kar S, Dutta A, et al. A multicenter prospective hospital-based surveillance to estimate the burden of rotavirus gastroenteritis in children less than five years of age in India. Vaccine. 2014;32(1):A13–A19. doi: 10.1016/j.vaccine.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Reviews in Medical Virology. 2005;15(1):29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- Sanz JC, Barbas JF, Lasheras MD, Jimenez M, Ramos B, Sanchez-Fauquier A. Detection of a rotavirus G9P[8] outbreak causing gastroenteritis in a geriatric nursing home. Enfermedades Infecciosas y Microbiología Clínica. 2009;27(4):219–221. doi: 10.1016/j.eimc.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Saudy N, Elshabrawy WO, Megahed A, Foad MF, Mohamed AF. Genotyping and clinicoepidemiological characterization of rotavirus acute gastroenteritis in Egyptian children. Polish Journal of Microbiology. 2017;65(4):433–442. doi: 10.5604/17331331.1227669. [DOI] [PubMed] [Google Scholar]
- Steele AD, Ivanoff B. Rotavirus strains circulating in Africa during 1996–1999: Emergence of G9 strains and P[6] strains. Vaccine. 2003;21(5–6):361–367. doi: 10.1016/s0264-410x(02)00616-3. [DOI] [PubMed] [Google Scholar]
- Tagbo BN, Chukwubike C, Mwenda JM, Seheri ML, Armah G, Mphahlele JM, et al. Molecular characterization of rotavirus strains circulating in Enugu Nigeria: 2011 to 2016. World Journal of Vaccines. 2019;9:22–36. [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, et al. Global, regional, and national estimates of rotavirus mortality in children %3c 5 years of age, 2000–2013. Clinical Infectious Diseases. 2016;62:96–105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- Than VT, Kim W. Prevalence of rotavirus genotypes in South Korea in 1989–2009: Implications for a nationwide rotavirus vaccine program. Korean Journal of Pediatrics. 2013;56(11):465–473. doi: 10.3345/kjp.2013.56.11.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, et al. Rotavirus efficacy and safety trial (REST) study team. Safety and efficacy of a pentavalent human–bovine (WC3) reassortant rotavirus vaccine. The New England Journal of Medicine. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- Villena C, El-Senousy WM, Abad FX, Pintó RM, Bosch A. Group A rotavirus in sewage samples from Barcelona and Cairo: Emergence of unusual genotypes. Applied and Environmental Microbiology. 2003;69:3919–3923. doi: 10.1128/AEM.69.7.3919-3923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub ES, Baggs J, Duffy J, Vellozzi C, Belongia EA, Irving S, et al. Risk of intussusceptions after monovalent rotavirus vaccination. The New England Journal of Medicine. 2014;370:513–519. doi: 10.1056/NEJMoa1311738. [DOI] [PubMed] [Google Scholar]
- Wen X, Cao D, Jones RW, Li J, Szu S, Hoshino Y. Construction and characterization of human rotavirus recombinant VP8* subunit parenteral vaccine candidates. Vaccine. 2012;30(43):6121–6126. doi: 10.1016/j.vaccine.2012.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C, Healy K, Tate JE, Parashar UD, Bines J, Neuzil K, et al. Rotavirus vaccination and intussusception—science, surveillance, and safety: A review of evidence and recommendations for future research priorities in low and middle income countries. Human Vaccines & Immunotherapeutics. 2016;12(10):2580–2589. doi: 10.1080/21645515.2016.1197452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yih WK, Lieu TA, Kulldorff M, Martin D, McMahill-Walraven CN, Platt R, et al. Intussuception risk after rotavirus vaccination in U.S. Infants. The New England Journal of Medicine. 2014;370:503–512. doi: 10.1056/NEJMoa1303164. [DOI] [PubMed] [Google Scholar]
- Yu J, Lai S, Geng Q, Ye C, Zhang Z, Zheng Y, et al. Prevalence of rotavirus and rapid changes in circulating rotavirus strains among children with acute diarrhea in China, 2009–2015. Journal of Infection. 2019;78(1):66–74. doi: 10.1016/j.jinf.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller M, Patton JT, Heylen E, De Coster S, Ciarlet M, Ranst V, et al. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. Journal of Clinical Microbiology. 2012;50(3):966–976. doi: 10.1128/JCM.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Lv D, Wang S, Lin X, Bi Z, Wang H, et al. Continuous detection and genetic diversity of human rotavirus A in sewage in eastern China, 2013–2014. Virology Journal. 2016;13(1):153–160. doi: 10.1186/s12985-016-0609-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.