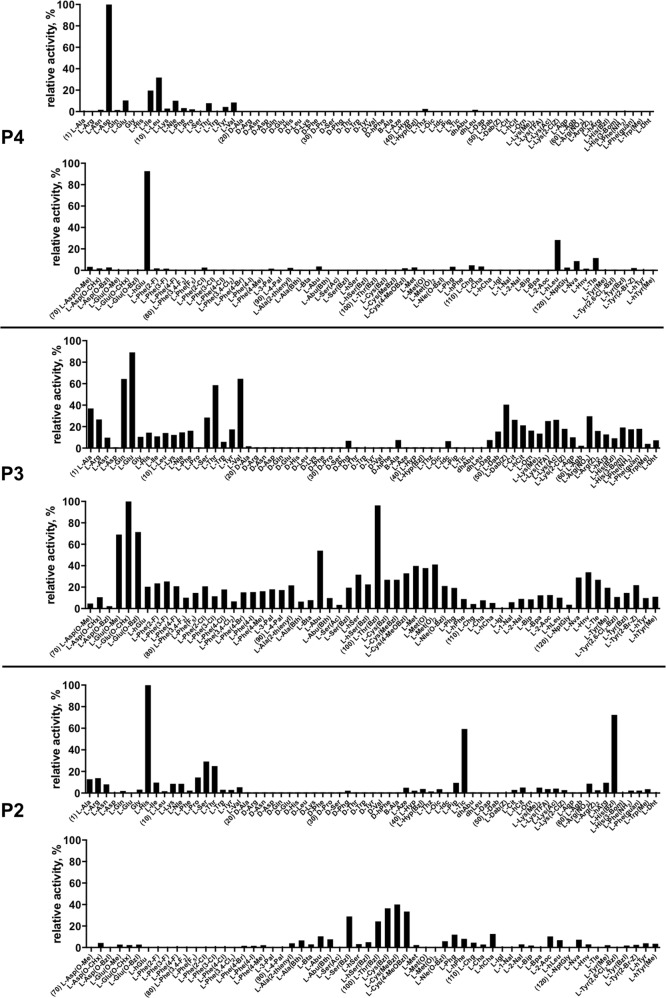
Fig. 1.
Human caspase-2 preferences at the P4, P3, and P2 positions. Human caspase-2 preferences were determined using three sub-libraries, Ac-P4-Mix-Mix-Asp-ACC, Ac-Mix-P3-Mix-Asp-ACC, Ac-Mix-Mix-P2-Asp-ACC, all containing natural and unnatural amino acids. The x axis shows abbreviated names of amino acids and the y axis displays relative activity presented as a percentage of the best-recognized amino acid. Standard deviations calculated from three screening were 15% of values shown in the figure. The order of the amino acids on the x axis corresponds to the order in Table S4. Every tenth amino acid is marked with a number