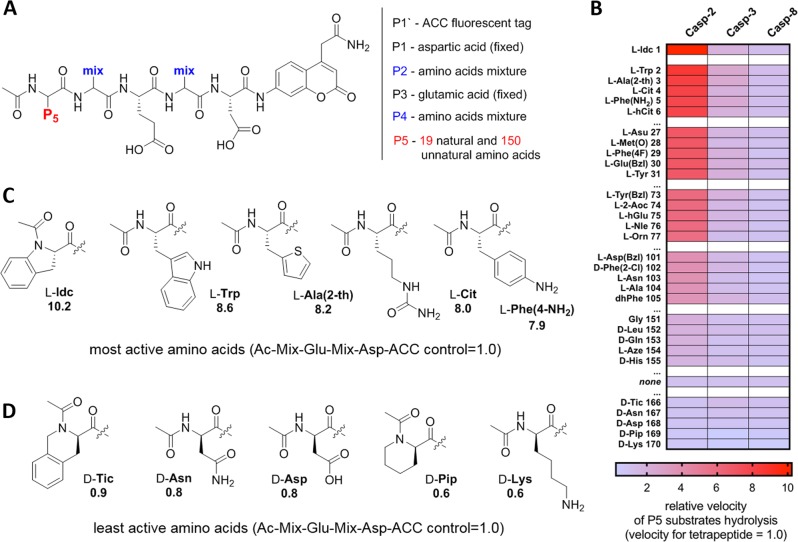
Fig. 2.
Analysis of caspase-2 P5 preferences. a General architecture of the P5 combinatorial fluorogenic substrate library, Ac-P5-Mix-Glu-Mix-Asp-ACC. The structure of this library allows for versatile screening of all caspases. b Substrate specificity of three apoptotic caspases at P5 position. The data are presented as a heat-map, where the velocity of substrate hydrolysis is expressed as ratio between P5 substrate (Ac-P5-Mix-Glu-Mix-Asp-ACC) and a tetrapeptide substrate lacking the P5 amino acid (Ac-Mix-Glu-Mix-Asp-ACC), which serves as a control. The results are sorted according to caspase-2 preferences, from the most to least active. The full P5 specificity profiles of these caspases can be found in Fig. S1. “none”—activity of tetrapeptide substrate lacking P5 amino acid. c, d The structures of the best (c) and worst (d) caspase-2 amino acids at P5 position. The numbers in c and d represent the ratio between cleavage rates of pentapeptides and Ac-Mix-Glu-Mix-Asp-ACC tetrapeptide, which served as a control