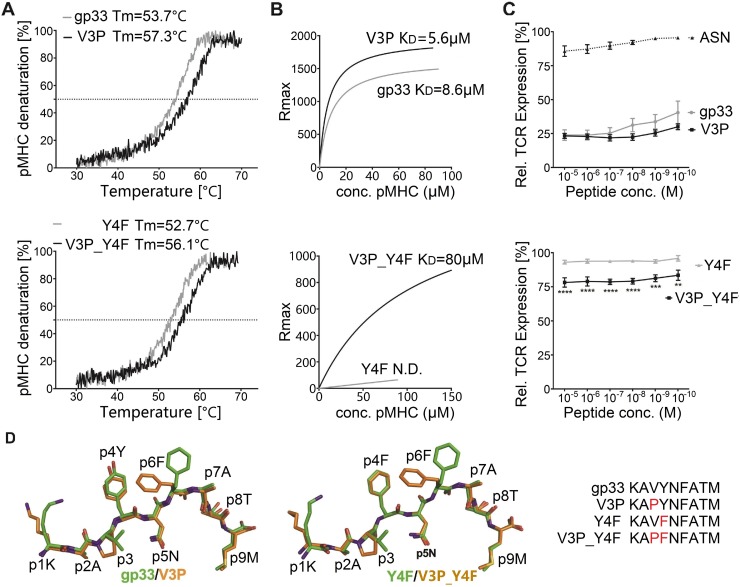
Fig 1. The p3P modification enhances pMHC stability without altering structural conformation, reestablishing TCR recognition.
A. The p3P modification increases pMHC stability. CD melting curves of H-2Db/gp33 and H-2Db/V3P (upper panel), and H-2Db/Y4F and H-2Db/V3P_Y4F (lower panel). Melting temperatures (Tm) corresponding to 50% protein denaturation are indicated. B. The soluble TCR P14 binds to the APL V3P_Y4F. In contrast to Y4F, V3P_Y4F is bound by P14. Binding affinity of the soluble TCR P14 to each pMHC was measured using SPR. KD values are indicated. C. The p3P modification increases TCR internalization. TCR downregulation was measured following exposure of P14 T cells to H-2Db in complex with each peptide at indicated concentrations on RMA cells. CD3+CD8+CD4- and Vα2+ cells were gated to quantify TCR internalization and p values calculated by using two-way Anova with Turkey’s multiple comparison test. **** represents p<0.0001; *** 0.0002 and ** 0.0018. The H-2Db-restricted Influenza-derived peptide ASNENMETM (ASN) was used as negative control. D. The p3P modification does not alter the conformation of the backbone of APLs compared to native counterparts. Superposition of the crystal structures of H-2Db/V3P and H-2Db/V3P_Y4F with H-2Db/gp33 and H-2Db/Y4F demonstrates that the p3P modification does not alter backbone conformations. Significant conformational changes are only observed for the side chains of peptide residues p1K and p6F following the p3P substitution.