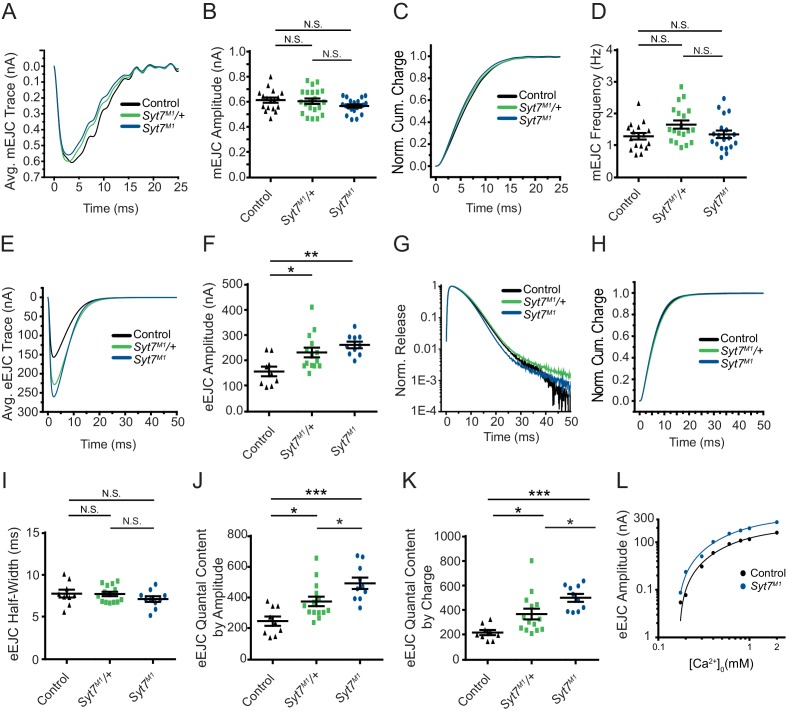
Figure 2. Syt7 mutants and Syt7/+ heterozygotes display enhanced neurotransmitter release.
(A) Average mEJC traces in control (black), Syt7M1/+ (green) and Syt7M1 mutants (blue). (B) Quantification of mean mEJC amplitude for the indicated genotypes (control: 0.62 ± 0.020 nA, n = 17; Syt7M1/+: 0.61 ± 0.021 nA, n = 21; Syt7M1: 0.57 ± 0.013 nA, n = 20). (C) Normalized cumulative mEJC charge for each genotype. (D) Quantification of mean mEJC frequency for the indicated genotypes (control: 1.30 ± 0.10 Hz, n = 17; Syt7M1/+: 1.66 ± 0.13 Hz, n = 19; Syt7M1: 1.36 ± 0.12 Hz, n = 19). (E) Average eEJC traces in control (black), Syt7M1/+ (green) and Syt7M1 (blue). (F) Quantification of mean eEJC amplitude for the indicated genotypes. (G) Average normalized responses for each genotype plotted on a semi-logarithmic graph to display release components. (H) Cumulative release normalized to the maximum response in 2 mM Ca2+ for each genotype. (I) Quantification of mean eEJC half-width in the indicated genotypes (control: 7.81 ± 0.47 ms, n = 9; Syt7M1/+: 7.77 ± 0.26 ms, n = 14; Syt7M1: 7.15 ± 0.34 ms, n = 10). (J) Quantification of evoked quantal content with mEJC amplitude for the indicated genotypes (control: 250.1 ± 30.58 SVs, n = 9; Syt7M1/+: 377.9 ± 31.13, n = 14; Syt7M1: 495.3 ± 36.75, n = 10). (K) Quantification of evoked quantal content with mEJC charge for the indicated genotypes (control: 221.3 ± 20.54 SVs, n = 9; Syt7M1/+: 371.6 ± 43.56, n = 14; Syt7M1: 503.6 ± 31.99, n = 10). (L) Log-log plot for eEJC amplitudes recorded in 0.175, 0.2, 0.3, 0.4, 0.6, 0.8, 1, and 2 mM extracellular [Ca2+] from control (black) and Syt7M1 mutants (blue), with a Hill fit for each genotype noted. Recordings were performed from 3rd instar segment A3 muscle 6. Extracellular [Ca2+] in E–K was 2 mM. Statistical significance was determined using one-way ANOVA (nonparametric) with post hoc Tukey’s multiple comparisons test. N.S. = no significant change. Error bars represent SEM.