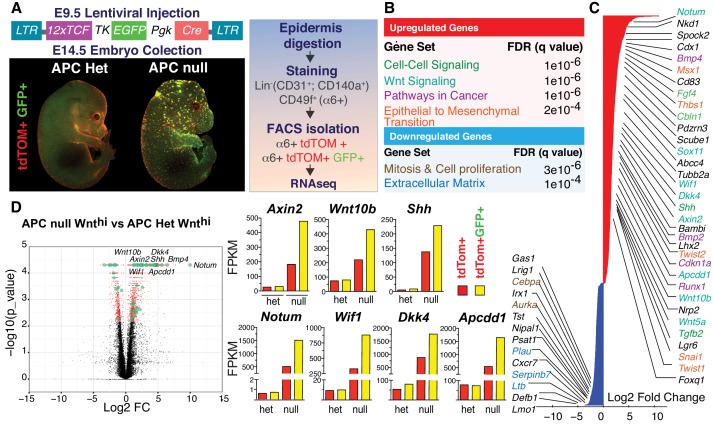
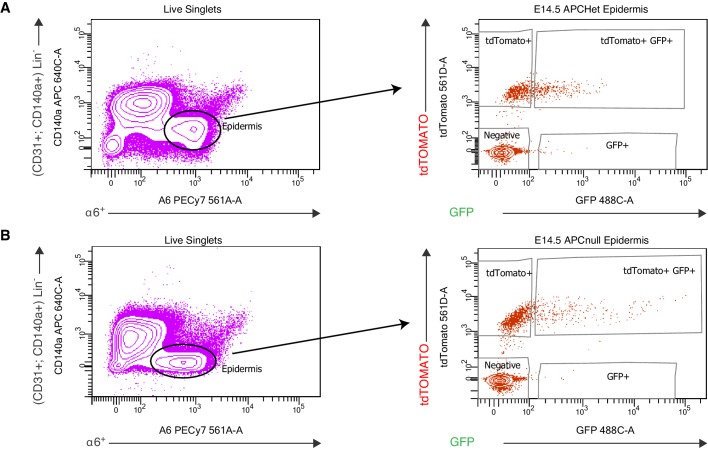
Figure 2. Teasing out a WNT-sensitive molecular signature based upon transcriptome profiling of skin progenitors possessing different WNT signaling levels.
(A) LV construct, epifluorescence imaging and FACS strategy for isolating WNT signaling (GFP+) and WNTlo skin progenitors from LV-transduced E14.5 Apc fl/+ and Apcfl/fl; R26fl-stop-fl-tdTOM embryos. (B) Gene set enrichment analysis (GSEA) of gene sets showing marked differential expression in WNT signaling progenitors from Apc-null vs Apc-het embryos. False discovery rate (FDR) q-values of enrichment are shown for each gene set. (C) Waterfall plot depicting genes markedly influenced (Log2 Fold Change ≥ 1.5, p<0.05) by APC status (color-coding according to B). (D) Volcano plot showing differentially regulated transcripts and WNT-reporter status.