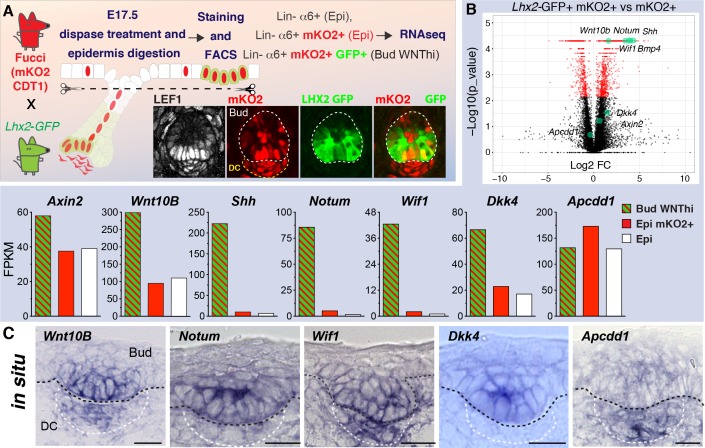
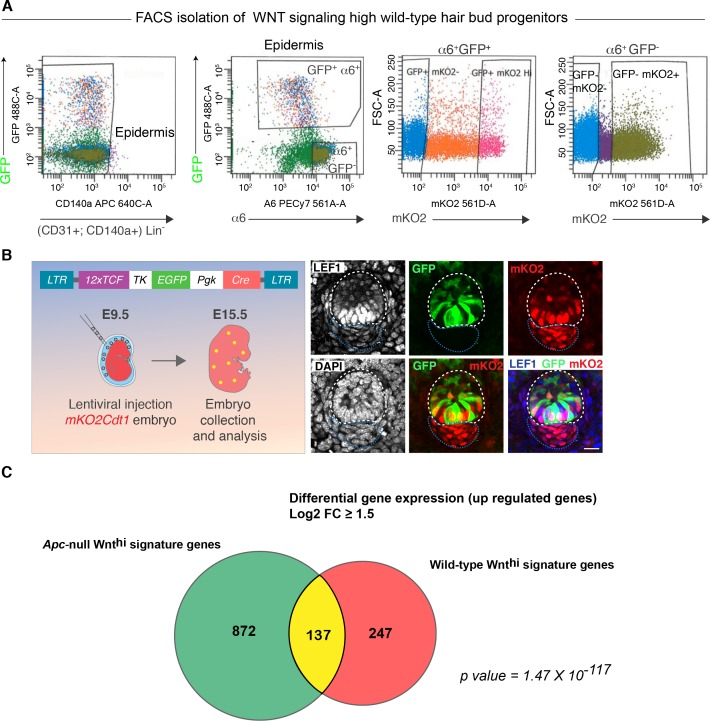
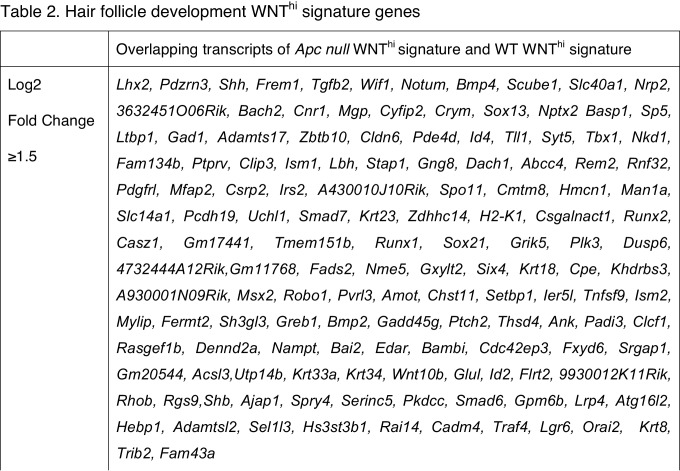
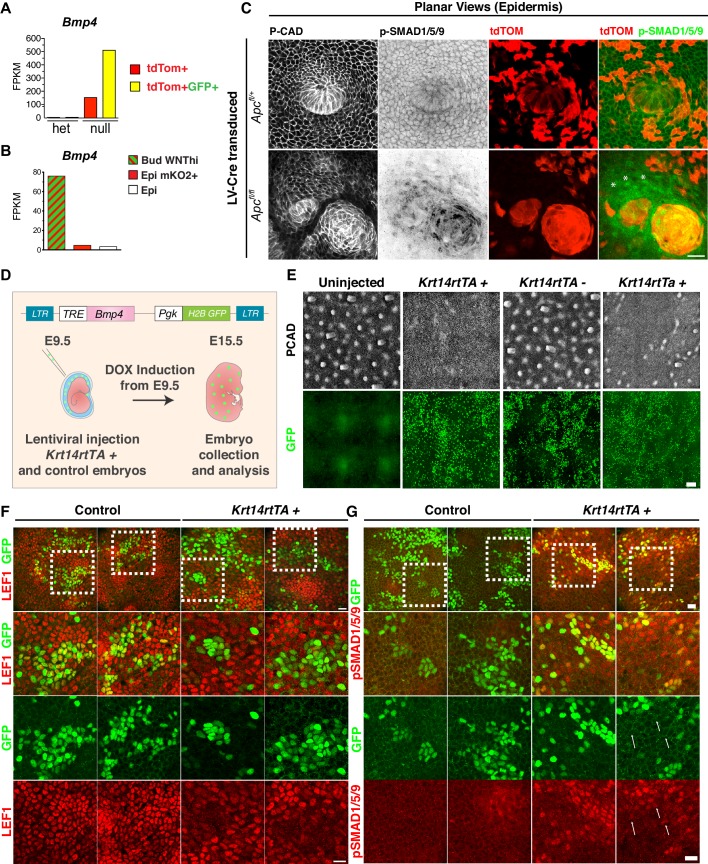
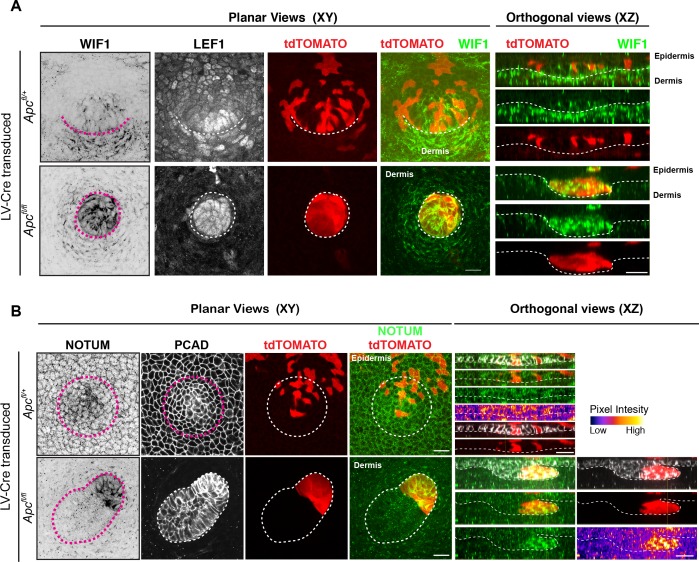
Figure 3. Wild-type WNT signaling progenitor cells express high levels of WNT inhibitors.
(A) Strategy used to isolate and profile slow-cycling basal progenitors from the epidermal fraction of dispase-treated, wild-type E17.5 skin, which contains epidermis and early hair placodes/buds. Note: LEF+ progenitors are simultaneously LHX2GFP+ and mKO2+. (B) Volcano plot and comparative expression profiling reveals that relative to their epidermal counterparts, wild-type basal placode/bud progenitors share strong signature similarities with Apc-null progenitors. Green dots denote previously reported WNT target genes. (C) In situ hybridizations showing that WNT signaling progenitor cells simultaneously express mRNAs for WNT activators and WNT inhibitors. Black dashed lines, epidermal-dermal boundary; white dashed lines demarcate the dermal condensate (DC). Scale bars, 10 μm.