Abstract
Soft tissue tumors with GLI1 gene fusions or amplifications have been recently described as a unique pathologic entity with established risk of malignancy. We herein expand these findings by investigating a cohort of 11 head and neck lesions with GLI1 alterations, including 8 from the tongue, for their clinicopathologic and molecular features. The tumors commonly affected males in their 30s (M:F ratio 2.7:1; range 1–65). Tumors showed a multinodular growth pattern, nested architecture separated by a delicate, arborizing vascular network, monotonous round to ovoid nuclei, and clear cytoplasm. Tumor protrusion into vascular spaces was common. Genetic alterations were investigated by FISH and/or targeted RNA sequencing. Seven tumors harbored GLI1 fusions with the following partners: ACTB (n=4), PTCH1 (n=2), or MALAT1 (n=1). The remaining four cases showed co-amplifications of GLI1 with CDK4 and MDM2 genes. Tumors were commonly positive for S100 protein and CD56. CDK4, MDM2 and STAT6 were positive in GLI1-amplified tumors. Two of six patients with available follow-up (one each with GLI1-amplification and PTCH1-GLI1 fusion) developed distant metastases. Both tumors showed a high mitotic index and tumor necrosis. The H&N region, particularly tongue, is a common location for GLI1-related mesenchymal tumors. Although a morphologic overlap was noted with the previously reported ‘pericytoma with t(7,12) translocation’, often occurring in the tongue, our findings expand the original findings, to include a more variable immunophenotype, propensity for late distant metastases, and alternative mechanisms of GLI1 oncogenic activation, such as various GLI1 fusion partners or GLI1 co-amplifications with MDM2 and CDK4 genes.
Keywords: GLI1 amplification, GLI1 fusion, ACTB, MALAT1, PTCH1, pericytoma
Introduction
GLI family zinc finger 1 (GLI1) belongs to the GLI gene family, encoding the GLI1 transcription factor, a key downstream effector of the hedgehog cascade. Aberrant activation of hedgehog signaling has been observed in a wide range of tumors, including basal cell carcinoma, gliomas, and pancreatic, colorectal, prostatic, lung and breast carcinomas (1).
In 2004, ACTB-GLI1 fusion was first described by Dahlen et al. in a so-called ‘pericytoma with t(7;12) translocation’, a rare but distinctive soft tissue neoplasm associated with a benign clinical course and purported pericytic differentiation based on its smooth muscle actin (SMA)-positive/S100-negative immunophenotype and ultrastructural characteristics (2, 3). However, several recent studies have demonstrated GLI1 genetic abnormalities in a group of soft tissue tumors with shared morphologic features but a more variable immunoprofile, which often includes S100 positivity, and propensity for malignant behavior, including regional and/or distant metastasis (4–9). Moreover, the genetic alterations encountered in these tumors included not only GLI1 fusions with ACTB gene, but also with other partners, such as MALAT1 and PTCH1, as well as high level GLI1 gene amplifications, often being co-amplified with nearby genes located on the 12q13.3-q15 region, such as DDIT3/CDK4/MDM2/STAT6 genes (6, 7).
Interestingly, the head and neck (HN) region, specifically the tongue, was the most common site of involvement for tumors defined originally as pericytoma with t(7;12), occurring in 3 of the 5 cases reported (3). In this study we investigate the clinicopathologic findings of a large cohort of HN mesenchymal neoplasms with various GLI1 genetic abnormalities and evaluate their pathogenetic relationship with the previously defined group of pericytic neoplasms.
Materials and Methods
Case selection and clinicopathologic review
The study was approved by the institutional review board. Eleven HN mesenchymal tumors harboring GLI1 alterations were retrieved from the archived pathology files and personal consultation files of the authors. Clinical features and outcomes, e.g. age, sex, site of the primary tumor, follow up period, recurrence, and distant metastasis, were gathered. All slides were centrally reviewed by BX and CRA to collect the following pathologic parameters: growth pattern, architecture, cytologic features, vascular pattern, vascular protrusion/invasion, mitotic index (per 10 high power fields, field diameter 0.55 mm), and tumor necrosis. Any unusual histologic features were documented. Available immunohistochemical stains were reviewed, including S100 protein, SOX10, CD56, keratin AE1/AE3, epithelial membrane antigen (EMA), desmin, SMA, STAT6, MDM2, and CDK4.
Detection of GLI1 genetic abnormalities
The underlying GLI1 gene molecular alterations were investigated by fluorescence in situ hybridization (n=10) and/or targeted RNA sequencing platform (n=2).
Fluorescence in situ hybridization (FISH)
FISH on interphase nuclei from formalin-fixed paraffin-embedded 4-μm sections was performed using custom designed probes of bacterial artificial chromosomes (BACs) flanking the target genes as previously described (supplementary table 1) (6, 7). Two hundred successive nuclei were examined for the presence of GLI1 alterations using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Waltham, MA). A positive FISH score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with an incomplete set of signals were omitted from the score. If GLI1-rearrangement was detected, subsequent FISH for PTCH1, MALAT1, and ACTB1 was performed to identify the fusion partners.
For cases with GLI1 gene amplification, the following FISH signal patterns were considered as positive results: 1) a homogeneously staining region; 2) double minutes; 3) ring chromosomes; and 4) multiple dot-like amplicons of various size, with at least 10:1 ratio to the centromeric 12 reference. Additional FISH assays for assessing MDM2 and CDK4 gene copy alterations were performed in GLI1-amplified tumors.
Targeted RNA sequencing (RNA-seq)
Details of the RNA-seq has been described previously (7, 10). In brief, RNA was extracted using the EXpressART FFPE Clear RNA Ready Kit (Amsbio, Cambridge, MA, US), and RNA-seq libraries prepared using the Illumina TruSight RNA fusion panel targeting 507 known fusion-related gene targets (Illumina, San Diego, CA, US). The tumor sample was sequenced using Illumina MiSEq V.3 platform. Sequenced reads passing filter in excess of 1.0 million indicate that the specimen quality passes the acceptable standard. Fusion gene analysis was performed using Illumina’s Local Run Manger (v. 1.3.0), with the STAR (version 2.3) and BOWTIE2 aligners, and the Manta and JAFFA fusion analysis, respectively. The mRNA expression level of genes of interests were evaluated and compared with those of other samples analyzed on the same targeted RNA-seq platform.
Statistics
All statistical analyses were performed using the SPSS software 24.0 (IBM Corporation, New York, NY, US). The clinicopathologic features were compared between GLI1-amplified and GLI1-translocated tumors using appropriate statistical tests, i.e. Fisher’s exact test for nonparametric variables and two-tailed Student’s t test for continuous variables. P values less than 0.05 were considered to be statistically significant.
Results
Clinical characteristics
Eleven patients with GLI1-altered mesenchymal tumors were included in this study. The clinicopathologic features are summarized in Table 1. Three cases (cases #1, #3, and #5) were retrospectively identified based on our genetic screening of unclassified tumors, being included in our previous publications first reporting this novel entity (6, 7), whereas the remaining 8 cases were prospectively recognized and collected based on their unique histologic features. The median age of diagnosis was 38 years (range: 1–65). There was a male predominance with a male to female ratio of 2.7:1. Two cases occurred in children, both presenting in the tongue and harboring GLI1-ACTB fusions, including an infant male with a congenital lesion and a 14-year-old boy. In fact, the tongue was the most common site of presentation, seen in all except 3 patients (73%). Other less commonly affected sites included submandibular gland, neck/submandibular gland, and soft tissues of the neck (one case each).
Table 1.
Head and neck GLI1-positive mesenchymal tumors: clinicopathologic, immunophenotypic and molecular characteristics.
| Molecular profile | Clinical characteristics | Pathologic features and immunoprofile | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GLI1 alteration (12q13.3) | CDK4 (12q14.1) | MDM2 (12q15) | Fusion Partner | Age | Sex | Primary site | LR/ DM | Status at last FU | FU period (months) | MI | Necrosis | S100 | SOX10 | CD56 | SMA | AE1/3 | EMA | STAT6 | CDK4 | MDM2 | |
| 1 | Amp. | Amp. | Amp. | 39 | M | Neck | Yes | AWD | 36 | 7 | Yes | + | - | +++ | +++ | ++ | - | - | + | + | |
| 2 | Amp. | Amp. | Amp. | 46 | F | Tongue | No | NED | 3 | 0 | No | + | - | +++ | ++ | - | - | - | +++ | + | |
| 3 | Amp. | Amp. | Amp. | 65 | M | Tongue | NA | NA | NA | 5 | No | - | - | ND | - | - | - | +++ | +++ | +++ | |
| 4 | Amp. | Amp. | Amp. | 60 | M | Tongue | NA | NA | NA | 11 | No | + | ND | ND | - | ND | ND | + | ND | ND | |
| 5 | Fusion | PTCH1 | 32 | F | Subm. | Yes | AWD | 104 | 6 | Yes | +++ | ND | ND | - | - | + | ND | ND | ND | ||
| 6 | Fusion | PTCH1 | 38 | M | Tongue | No | NED | 2 | 1 | No | ++ | - | +++ | - | - | ND | ND | ND | ND | ||
| 7 | Fusion | ACTB | 37 | M | Neck/subm. | No | NED | 30 | 0 | No | - | ND | +++ | + | - | ND | - | - | + | ||
| 8 | Fusion | ACTB | 1 | M | Tongue | No | NED | 2 | 4 | No | ND | ND | ND | - | ND | ND | ND | ND | ND | ||
| 9 | Fusion | ACTB | 28 | M | Tongue | NA | NA | NA | 0 | No | ++ | - | ND | + | - | - | ND | ND | ND | ||
| 10 | Fusion | ACTB | 14 | M | Tongue | NA | NA | NA | 0 | No | - | ND | ND | ND | - | ND | ND | ND | ND | ||
| 11 | Fusion | MALAT1 | 56 | F | Tongue | NA | NA | NA | 8 | No | - | - | ND | - | + | ND | ND | ND | ND | ||
FU: follow up; MI: mitotic index (per 10 high power fields); SMA: smooth muscle actin; AE1/3: cytokeratin AE1/AE3; EMA: epithelial membrane antigen; Amp.: Amplification; M: male; F: female; Subm.: submandibular gland; LR: local recurrence; DM: distant metastasis; AWD: alive with disease; NED: no evidence of disease; NA: not available; ND: not done; +++: diffuse and strong immunopositivity; ++: Patchy or multifocal immunopositivity; +: focal or weak immunopositivity; -: negative.
Pathologic features and immunophenotype
The typical histologic features seen in all cases included a multinodular or plexiform growth pattern at low magnification, with a distinctive nested architecture separated by a rich delicate arborizing vascular network. The constituent cells had a monomorphic cytomorphology, with uniform round to ovoid nuclei and often clear cytoplasm (Fig. 1). Protrusion of tumor nests into vascular spaces was a common finding, observed in all except 3 cases (75%). The mitotic activity was variable, ranging from 0 to 11 per 10 high power fields (median: 4).
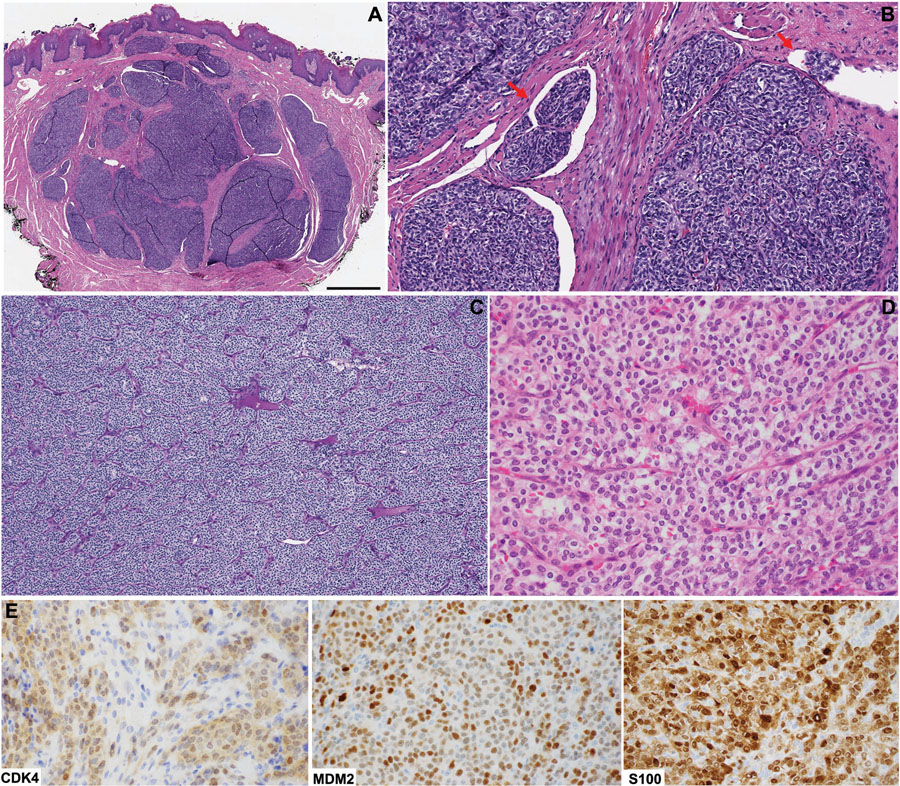
Figure 1. Head and neck mesenchymal tumor with GLI1 gene abnormalities: typical histologic features.
(A) A tongue tumor positive for GLI1-ACTB fusion showing the typical multilobulated/plexiform growth pattern at low power (scale bar: 1 cm). (B) Tumor nests protruding within vascular channels (red arrows) is a common histologic finding. (C) Solid architecture divided in subtle nests of various sizes by rich delicate arborizing vascular network. (D) At high power, the tumors cells have monotonous round to ovoid nuclei and moderate amount of clear to eosinophilic cytoplasm. The underlying molecular alterations may vary, including ACTB-GLI1 fusion (A/B), PTCH1-GLI1 fusion (C), and GLI1 amplification (D). (E) By immunohistochemistry, the GLI1-amplified tumor shown in panel D is positive for CDK4, MDM2, and S100 protein.
Other uncommon microscopic findings that might present infrequently and focally in these tumors included prominent cytoplasmic vacuolization resembling lipoblasts/pseudolipoblasts (n=1); distinct spindling and fascicular growth (n=1); abundant eosinophilic granular cytoplasm (n=1); pseudorosettes (n=2), tumor necrosis (n=2); prominent fibrous septa (n=3); microcystic/pseudoalveolar/pseudoglandular change (n=2); cord like arrangement of tumor cells within a prominent myxoid stroma (n=2); and stromal hyalinization with multiple hyaline globules (n=1, Fig. 2).
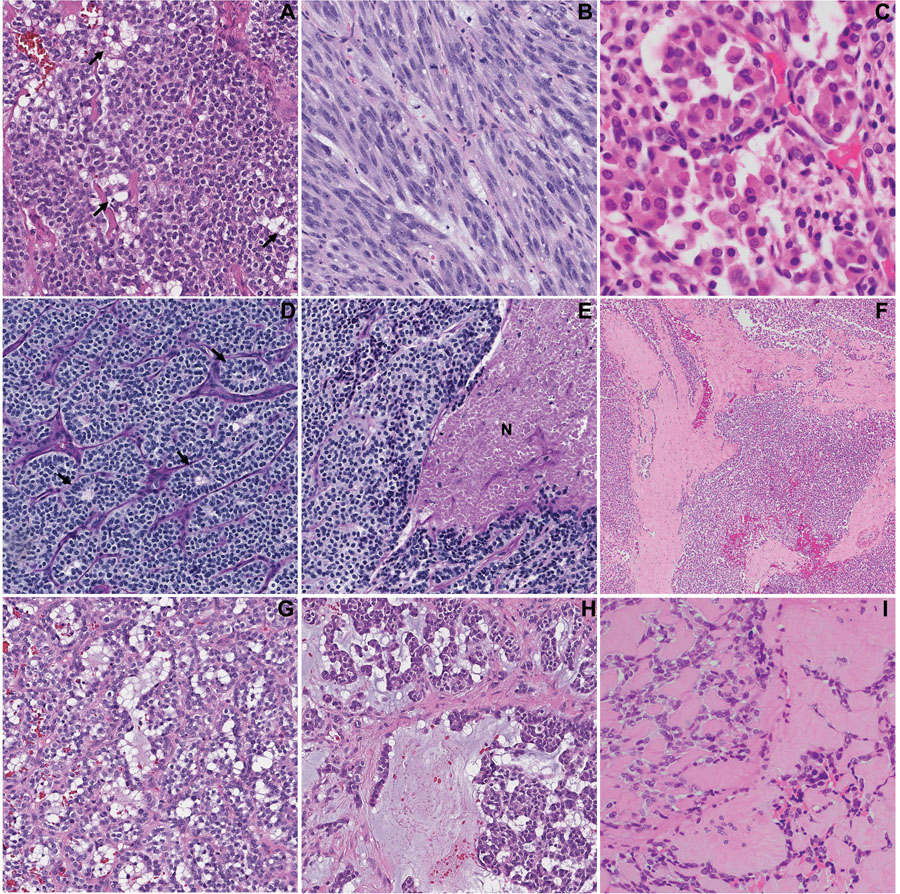
Figure 2. Uncommon histologic features of head and neck mesenchymal tumor with GLI1 alterations:
(A) Pseudolipoblast appearance with prominent cytoplasmic vacuolization; (B) prominent spindling and fascicular arrangement; (C) nested to alveolar growth, with cells showing abundant, densely eosinophilic cytoplasm; (D) pseudorossettes (arrows); (E) tumor necrosis (N), (F) thick fibrous bands; (G) microcystic/pseudoglandular pattern; (H) cord arrangement within prominent myxoid stroma; and (I) prominent hyalinized stroma and hyaline globules. These features may present infrequently and focally within a GLI1-altered tumor.
Immunohistochemical results are summarized in Table 1. Tumors were often positive for S100 protein (6/10, 60%) and showed strong and diffuse CD56 staining (4/4, 100%). Additional SMA, keratin AE1/AE3 and EMA immunopositivity was observed in 40% (4/10), 22% (2/9) and 20% (1/5) of cases, respectively. Other markers, including SOX10, CD31, CD34, ERG, chromogranin, synaptophysin, CD99 and desmin were consistently negative.
Molecular findings and clinicopathologic correlates
GLI1 alterations were confirmed in all tumors using FISH and/or RNA-seq platforms. Seven tumors harbored GLI1 fusions with the following partners: ACTB (n=4), PTCH1 (n=2) or MALAT1 (n=1). The remaining four cases showed co-amplifications of GLI1 with CDK4 and MDM2 genes (Fig. 3). GLI1-amplified tumors often demonstrated overexpression of CDK4 (3/3, 100%), MDM2 (3/3, 100%), and STAT6 (3/5, 60%) detected by immunohistochemistry.
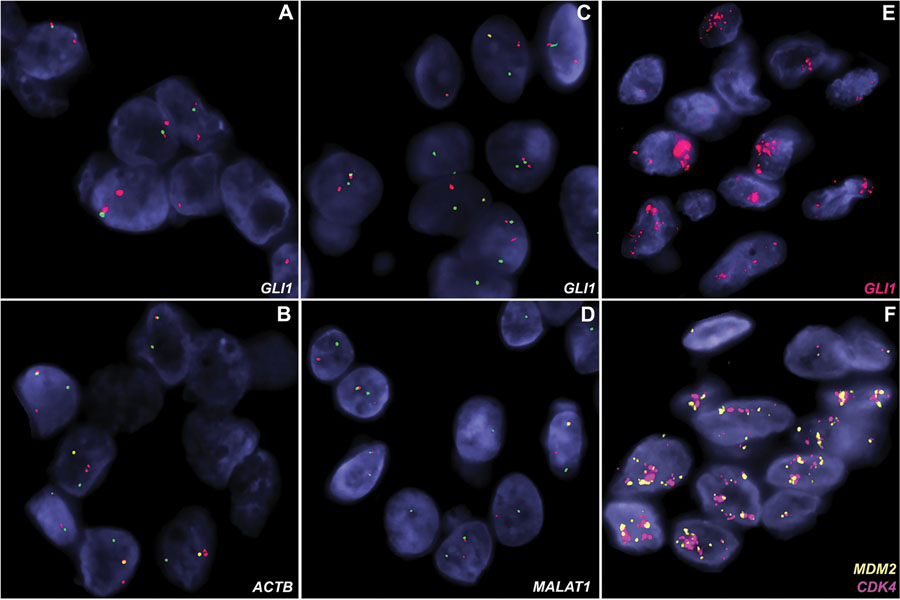
Figure 3. GLI1 alterations demonstrated using fluorescence in situ hybridization (FISH).
(A-B) A tumor with ACTB-GLI1 fusion showing GLI1 (A) and ACTB (B) break-apart signals (red, centromeric; green, telomeric). (C-D) A tumor with MALAT1-GLI1 fusion showing rearrangements for both GLI1 (C) and MALAT1 genes with split-apart signals (red, centromeric; green, telomeric). (E-F) A lesion contains high level co-amplifications of GLI1 (red, panel E), MDM2 (orange, panel F), and CDK4 (red, panel F) genes.
In our cohort, GLI1-amplified tumors were associated with significant older age at presentation (median: 53, range: 39–65) compared with GLI1-translocated tumors (median: 32, range 1–56, two-tailed Student’s t test, p=0.047). Other clinicopathologic parameters (e.g. gender, mitotic index, necrosis, and immunophenotype) did not differ significantly between the two molecular groups.
Clinical outcome highlighting the potential for distant metastasis
Six patients had clinical follow up data available with a median follow up of 16.5 months (range: 2–104 months). Two patients (one with GLI1-amplification and the other with PTCH1-GLI1 fusion) developed local recurrence and distant metastases, at 6 and 83 months, respectively. The metastatic sites were lung (n=2), bone (n=1), soft tissue (n=1) and brain (n=1). Both tumors showed a high mitotic index of 6 to 7 per 10 high power fields and tumor necrosis. The distant metastasis was available for review in one case (case #1), and showed typical histologic features as illustrated in Fig. 1, as well as areas of prominent spindling, elevated mitotic index (40 per 10 high power fields) and tumor necrosis. The remaining four cases were disease free and recurrence free at the last follow up.
Discussion
GLI1 genetic abnormalities, including GLI1 gene fusions and GLI1 amplifications, have been recently reported in a subset of soft tissue tumors with distinctive epithelioid nested phenotype and propensity for locoregional and distant metastasis. Although GLI1 fusion positive tumors are often S100 protein positive, the immunoprofile of tumors showing co-amplification of GLI1, MDM2 and CKD4 genes has been more variable and not indicative of a specific line of differentiation. Particularly intriguing is the pathogenetic relationship between this group of soft tissue tumors and the so-called ‘pericytoma with t(7;12) translocation’, a lesion that often occurs in the tongue and shows similar morphologic features, but often expresses SMA and is associated with a benign clinical course. In order to investigate this further, we carried out a detailed clinicopathologic and molecular analysis of a large cohort of cases presenting in the head and neck region harboring GLI1 genetic abnormalities.
Among the 35 cases of mesenchymal neoplasms with GLI1 alterations reported to date (2–9) (Table 2), 14 (40%) occurred within the HN region (2, 3, 6, 7). Strikingly, GLI1-positive HN tumors have a predilection for the tongue, with 73% of cases in the current cohort and 79% (11/14) of all reported cases occurring at this site. Other rare sites include submandibular gland and soft tissue of the neck. Although there is a male predominance in the current cohort, the overall gender distribution for all sites is quite balanced in the reported cases (2–9). Moreover, age range at presentation among the 35 patients reported is quite wide (range 1–79), but most commonly affect patients in their 30s (median, 37). Similarly, in the HN cohort, the median age at diagnosis is 38 (range 1–65). One outlier case was that of an ACTB-GLI1 translocated tumor of the tongue occurring in a 1-year-old male. The tumor was diagnosed since birth and represents the only congenital/infantile case reported to date.
Table 2.
Literature review of tumors with GLI1 gene alternations involving head and neck and other sites
| Age/sex | Site | Outcome (FU months) | MI | Necrosis | GLI1 alteration | S100 | SMA | AE1/3 | |
|---|---|---|---|---|---|---|---|---|---|
| Head and neck region | |||||||||
| Dahlen (2, 3) | 27/F | Tongue | NED (60) | NA | NA | ACTB-GLI1 | - | + | - |
| 11/M | Tongue | NED (22) | NA | NA | ACTB-GLI1 | - | + | - | |
| 12/F | Tongue | NED (120) | NA | NA | ACTB-GLI1 | - | + | - | |
| Antonescu (7) * | 32/F | Subm | LR/DM/AWD (104) | 6 | Yes | PTCH1-GLI1 | + | - | - |
| * | 38/M | Tongue | NED (2) | 1 | No | PTCH1-GLI1 | + | - | - |
| 37/M | Neck/Subm | NED (30) | 0 | No | ACTB-GLI1 | - | + | - | |
| 1/M | Tongue | NED (2) | 4 | No | ACTB-GLI1 | NA | - | NA | |
| 28/M | Tongue | NA | 0 | No | ACTB-GLI1 | + | + | - | |
| 14/M | Tongue | NA | 0 | No | ACTB-GLI1 | - | NA | - | |
| 56/F | Tongue | NA | 8 | No | MALAT1-GLI1 | - | - | + | |
| 46/F | Tongue | NED (3) | 0 | No | GLI1 amp | + | + | - | |
| 60/M | Tongue | NA | 11 | No | GLI1 amp | + | - | NA | |
| Agaram (6) * | 39/M | Neck | LR/DM/AWD (36) | 7 | Yes | GLI1 amp | + | + | + |
| 65/M | Tongue | NA | 5 | No | GLI1 amp | - | - | - | |
| Other sites | |||||||||
| Koh (4) | 11/F | Ovary | NA | NA | Yes | ACTB-GLI1 | + | - | - |
| Kerr (5) | 57/F | Tibia | DM/AWD (27) | 5 | Yes | ACTB-GLI1 | - | + | - |
| 62/M | Scapula | DM/AWD (180) | 1 | No | ACTB-GLI1 | - | + | - | |
| 41/F | Ovary | NED (14) | 1 | Yes | ACTB-GLI1 | + | + | + | |
| Antonescu (7) | 20/M | Thigh | NA | NA | No | ACTB-GLI1 | + | - | - |
| 16/M | Spine | NA | NA | No | MALAT1-GLI1 | + | - | - | |
| 30/F | Foot | LR/AWD (21) | NA | No | ACTB-GLI1 | + | - | - | |
| 79/F | RP | RR (NA) | NA | No | ACTB-GLI1 | - | - | - | |
| 38/F | Chest wall | NA | NA | No | ACTB-GLI1 | - | - | + | |
| Castro (8) | 8/F | Stomach | NED (6) | NA | Yes | ACTB-GLI1 | - | NA | - |
| Bridge (9) | 67/M | Bone | NED (204) | NA | NA | ACTB-GLI1 | - | + | - |
| Dahlen (2, 3) | 61/M | Calf | DOC (60) | NA | NA | ACTB-GLI1 | - | + | - |
| 65/F | Stomach | NED (24) | NA | NA | ACTB-GLI1 | - | + | - | |
| Agaram (6) | 4/F | Shoulder | NA | 5 | No | GLI1 amp | + | - | - |
| 10/M | Finger | NA | 15 | No | GLI1 amp | - | - | - | |
| 17/M | Thigh | NA | 5 | No | GLI1 amp | - | - | - | |
| 23/F | Thigh | NED (36) | 2 | No | GLI1 amp | - | - | + | |
| 26/F | Lung | NA | 4 | No | GLI1 amp | + | - | - | |
| 51/F | Back | LR/NED (16) | >25 | Yes | GLI1 amp | + | - | - | |
| 54/F | Elbow | NA | 15 | No | GLI1 amp | + | + | - | |
| 60/M | Forearm | NA | 10 | No | GLI1 amp | - | - | - | |
Current study; FU: follow up; MI: mitotic index (per 10 high power fields); SMA: smooth muscle actin; AE1/3: cytokeratin AE1/AE3; subm: submandibular gland; RP: retroperitoneum; NED: no evidence of disease; LR: local recurrence; DM: distant metastasis; RR: regional recurrence; AWD: alive with disease; NA: not available; DOC: dead of other cause; amp: amplification; +: positive; -: negative.
GLI1-positive tumors, including those originating in the HN region, share several characteristic microscopic features, such as a multinodular growth pattern, distinctive nested architecture separated by a delicate but rich arborizing capillary network, monomorphic cytomorphology with clear or pale eosinophilic cytoplasm and round to ovoid nuclei, and frequent protrusions of tumor nests into dilated vascular spaces. Recognizing these pathognomonic microscopic features (as illustrated in Fig. 1) may guide the pathologist towards the appropriate diagnosis and subsequent molecular testing if needed.
In the initial series of ‘pericytoma with t(7;12)’, the five reported tumors harboring ACTB-GLI1 fusions showed a consistent immunoprofile of SMA and laminin positivity, without S100 protein or keratin AE1/AE3 reactivity (3). Based on this immunoprofile and ultrastructural findings of prominent external lamina and subplasmalemmal density, the authors postulated that the tumor showed pericytic differentiation (3). More recently, however, it has become apparent that the immunoprofile of these tumors is much more variable and not indicative of a definitive line of differentiation (4–7). In the 36 reported cases to date, which include cases harboring both GLI1 fusions and amplifications, the cumulative frequency of immunopositivity for SMA, S100 protein, and keratin AE1/AE3 was 42% (14/33), 44% (15/34 cases), and 15% (5/33) respectively (Table 2). Additionally, tumors are frequently positive for CD56 in a diffuse fashion, but negative for neuroendocrine (synaptophysin and chromogranin), myoepithelial (GFAP and calponin), vascular (CD31 and ERG), SOX10, and muscle (desmin, myogenin and myoD1) markers. Additionally, the common presence of S100 protein expression and the occasional presence of keratin positivity would seem to argue against (myo)pericytic origin. Moreover, true pericytic tumors, such as glomus tumors or myopericytomas, which are positive for SMA, muscle specific actin (MSA), and h-caldesmon, and negative for keratin, S100 protein and desmin (11), harbor distinct genetic alterations (12, 13).
Within the HN region, as the tumor may occur in children and in major salivary gland, the differential diagnoses include primary salivary gland epithelial tumors, in particular sialoblastoma and myoepithelial carcinoma. Sialoblastoma is a rare salivary gland neoplasm of infancy which commonly contains organoid nests of primitive tumor cells with round to ovoid nucleoli (14). However, it lacks the clear cell features and the delicate vascular network of tumors with GLI1-alteration and may show areas of ductal differentiation and/or adenoid cystic carcinoma-like pattern. The molecular pathogenesis of sialoblastoma remains unknown. Of note, no GLI1 gene alterations were identified in one sialoblastoma case tested by FISH (data not shown). Recently, ACTB-GLI1 fusion has been also reported in gastroblastoma, a distinctive keratin-positive biphasic neoplasm affecting the stomach in children and young adults (15).
Other possible diagnostic mimics includes myoepithelioma and myoepithelial carcinoma, due to overlapping multilobulated growth, nested epithelioid morphology, clear cytoplasm (Fig. 2H) and S100/keratin immunopositivity (16). However, myoepitheliomas are well-circumscribed/encapsulated, in contrast with the infiltrative/multinodular growth of our cohort. Useful diagnostic clues for tumors with GLI1-alteration includes the rich vascular network separating the nested architecture, tumor bulging into vascular spaces and the absence of SOX10, GFAP, and calponin immunopositivity. Identifying areas with classic features of GLI1-altered tumors should trigger subsequent molecular confirmation as GLI1 alteration has not been reported in myoepithelial carcinoma of salivary gland (17). Moreover, distinct from pleomorphic adenoma, which shows a biphasic phenotype with both duct and myoepithelial differentiation, all our GLI1-altered tumors were composed of one cell type.
In view of the nested growth pattern and diffuse CD56 immunopositivity, paraganglioma in the HN region may be considered. Absence of immunoexpression of synaptophysin and chromogranin excludes such diagnosis.
Ectomesenchymal chondromyxoid tumor is another tumor included in the differential diagnosis of GLI1-altered tumors. It is a rare mesenchymal neoplasm characterized by a striking predilection for the anterior dorsal tongue, has a multilobulated growth pattern, and immunopositivity of S100, CD56, cytokeratin, and SMA (18, 19). However, ectomesenchymal chondromyxoid tumor typically shows a spindle cytomorphology, abundant myxoid stroma, lacking arborizing capillary network, and may show expression for other myoepithelial markers (e.g. calponin and GFAP). At the molecular level, tumors harbor a characteristic RREB1-MKL2 fusion, thus molecular testing can be used in challenging cases (18).
As observed in our prior study (6), tumors with GLI1 amplification commonly show co-amplification of nearby genes, including STAT6, CDK4 and/or MDM2, resulting in overexpression of STAT6, CDK4, and/or MDM2 protein, detectable by immunohistochemistry. Based on this immunoprofile, potential diagnostic pitfalls include a solitary fibrous tumor and a dedifferentiated liposarcoma, due to overlapping positivity for STAT6 and overexpression/gene amplification by FISH of MDM2 or CDK4, respectively. Both entities have been reported in the HN region (20, 21). However, GLI1-positive tumors typically lack CD34 positivity or areas of well-differentiated liposarcoma and have distinctive morphologic features.
In the original report of ‘pericytoma with t(7;12)’, patients followed a benign clinical course without recurrence and/or metastasis (2, 3). However, the recent series including tumors with either GLI1 gene fusions or GLI1 amplifications have documented the potential of these tumors for malignant behavior, including local recurrence and metastasis. Among the 20 reported cases with available follow-up data, four (20%) developed local recurrence, one (5%) regional lymph node metastasis and four (20%) distant metastasis. Three of the 4 cases with distant metastasis (5–7), had a high mitotic index of ≥5 per 10 high power fields (HPFs) and tumor necrosis. The fourth tumor reported by Kerr et al. (5) showed a mitotic index of 1 per 10 HPFs and lacked tumor necrosis, but developed distant metastasis to the lung 2 years after the initial resection. In contrast, only one (14%) of seven and three (30%) of ten tumors without distant metastasis showed elevated mitotic rate of ≥5/10 HPFs and necrosis respectively. The underlying molecular alterations of these four tumors with distant metastasis were diverse, with ACTB-GLI1 fusion in two, PTCH1-GLI1 fusion in one, and GLI1 amplification in the fourth case.
Tumors originated from the HN region followed a similar clinical course: two (22%) of nine reported cases developed local recurrence and distant metastasis (2–9). These 2 cases originated outside the tongue, one from the submandibular gland and the other from the neck area. Both tumors had an elevated mitotic index (6 to 7/10 high power fields) and tumor necrosis. One harbored GLI1 amplification, whereas the other contained a PTCH1-GLI1 fusion. All six patients with tongue tumors and available clinical follow-up were disease free at their last visit. Although the current evidence seems to suggest that elevated mitotic activity, tumor necrosis and an anatomic location outside of oral tongue may be associated with aggressive behaviors, larger series with longer follow-up are needed to determine if tumors with GLI1-alteration can be graded based on these pathologic features.
GLI1 oncogenic activation in HN tumors, similar to other soft tissue sites, may occur through gene fusion with various partners (ACTB, PTCH1, or MALAT1) or through amplification of GLI1 gene on chromosome 12q13.3 with often co-amplification of nearby genes, e.g. STAT6 (12q13.3), DDIT3 (12q13.3), CDK4 (12q14.1) and MDM2 (12q.15). In this study, GLI1-amplified tumors occurring in the HN region were associated with a significantly older age at presentation compared to those positive for GLI fusion. However, pooled data from the 35 published cases showed no significant age difference in these two molecular groups (two-tailed Student’s t test, p=0.736). The median age at diagnosis (range) for GLI1-amplified and GLI1-translocated tumors is 42.5 (4–65) and 32 (1–79), respectively. Additionally, other clinical, histologic, immunophenotypic features, and outcome do not differ between the two molecular groups. The cumulative evidence suggests that these tumors may be grouped under one umbrella as ‘sarcoma of unknown lineage with GLI1 alteration’.
Several inhibitors targeting the hedgehog pathway, including GLI inhibitors (e.g. arsenic trioxide, pirfenidone, and imiquimod) are currently approved by the U.S. Food and Drug Administration (FDA) or are available in various clinical trials to treat leukemia, basal cell carcinoma, and other types of carcinoma (1). Identifying GLI1 alteration and oncogenic activation in this unique tumor entity may therefore allow the patients, especially those with distant metastasis, to gain access to hedgehog or GLI1 targeted therapies.
In summary, we have reported 11 mesenchymal neoplasms with GLI1-gene alterations occurring in the HN region, the largest cohort of this entity to date. Our cohort showed that in the HN area, there is a striking predilection for oral tongue location. Similar to other soft tissue locations, HN GLI1-positive mesenchymal neoplasms are characterized by distinctive histologic features, including multinodular growth, nested architecture of epithelioid cells with clear cytoplasm and uniform ovoid to round nuclei, and a delicate arborizing capillary network. The findings of the current study, together with other recent series, expand our knowledge of the variable immunophenotype, molecular pathogenesis and clinical behaviors of these tumors, which were initially reported under the designation of “pericytoma with t(7;12) translocation”. The mechanism of GLI1 oncogenic activation in these tumors occurs through either gene fusions with various partners or through high level amplifications of the GLI1 gene locus, which often results in co-amplification of other neighboring genes at 12q13.3-q15 region. The emerging evidence suggests that these lesions should be regarded at least as low grade sarcoma, with a 20% rate of local recurrence, regional and distant metastases, especially if associated with worrisome histologic features. Their malignant behavior appears to be independent to the type of GLI1 genetic alteration.
Supplementary Material
Acknowledgments
Source of Funding
Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30 CA008748 (BX, CRA), P50 CA217694 (CRA), and P50 CA140146-01 (CRA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest
The authors have disclosed that they have no significant relationships with, or financial interest in any commercial companies pertaining to this article.
References
- 1.Didiasova M, Schaefer L, Wygrecka M. Targeting GLI Transcription Factors in Cancer. Molecules (Basel, Switzerland). 2018;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahlen A, Mertens F, Mandahl N, et al. Molecular genetic characterization of the genomic ACTB-GLI fusion in pericytoma with t(7;12). Biochemical and biophysical research communications. 2004;325:1318–1323. [DOI] [PubMed] [Google Scholar]
- 3.Dahlen A, Fletcher CD, Mertens F, et al. Activation of the GLI oncogene through fusion with the beta-actin gene (ACTB) in a group of distinctive pericytic neoplasms: pericytoma with t(7;12). The American journal of pathology. 2004;164:1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh NWC, Seow WY, Lee YT, et al. Pericytoma With t(7;12): The First Ovarian Case Reported and a Review of the Literature. International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists. 2019;38:479–484. [DOI] [PubMed] [Google Scholar]
- 5.Kerr DA, Pinto A, Subhawong TK, et al. Pericytoma With t(7;12) and ACTB-GLI1 Fusion: Reevaluation of an Unusual Entity and its Relationship to the Spectrum of GLI1 Fusion-related Neoplasms. The American journal of surgical pathology. 2019;43:1682–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agaram NP, Zhang L, Sung YS, et al. GLI1-amplifications expand the spectrum of soft tissue neoplasms defined by GLI1 gene fusions. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2019;32:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonescu CR, Agaram NP, Sung YS, et al. A Distinct Malignant Epithelioid Neoplasm With GLI1 Gene Rearrangements, Frequent S100 Protein Expression, and Metastatic Potential: Expanding the Spectrum of Pathologic Entities With ACTB/MALAT1/PTCH1-GLI1 Fusions. The American journal of surgical pathology. 2018;42:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro E, Cortes-Santiago N, Ferguson LM, et al. Translocation t(7;12) as the sole chromosomal abnormality resulting in ACTB-GLI1 fusion in pediatric gastric pericytoma. Human pathology. 2016;53:137–141. [DOI] [PubMed] [Google Scholar]
- 9.Bridge JA, Sanders K, Huang D, et al. Pericytoma with t(7;12) and ACTB-GLI1 fusion arising in bone. Human pathology. 2012;43:1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson BC, Hornick JL, Fletcher CDM, et al. Dermatofibrosarcoma protuberans with a novel COL6A3-PDGFD fusion gene and apparent predilection for breast. Genes, chromosomes & cancer. 2018;57:437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher CD, Bridge JA, Hogendoorm PCW, et al. WHO classification of tumours of soft tissue and bone. Lyon: International Agency for Research on Cancer; 2013. [Google Scholar]
- 12.Mosquera JM, Sboner A, Zhang L, et al. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes, chromosomes & cancer. 2013;52:1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung YP, Fletcher CDM. Myopericytomatosis: Clinicopathologic Analysis of 11 Cases With Molecular Identification of Recurrent PDGFRB Alterations in Myopericytomatosis and Myopericytoma. The American journal of surgical pathology. 2017;41:1034–1044. [DOI] [PubMed] [Google Scholar]
- 14.El-Naggar AK, Chan JKC, Grandis JR, et al. World Health Organization Classification of Tumours: pathology and genetics of head and neck tumours (4th edition). Lyon: International Agency for Research on Cancer (IARC); 2017. [Google Scholar]
- 15.Graham RP, Nair AA, Davila JI, et al. Gastroblastoma harbors a recurrent somatic MALAT1-GLI1 fusion gene. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2017;30:1443–1452. [DOI] [PubMed] [Google Scholar]
- 16.Xu B, Mneimneh W, Torrence DE, et al. Misinterpreted Myoepithelial Carcinoma of Salivary Gland: A Challenging and Potentially Significant Pitfall. The American journal of surgical pathology. 2019;43:601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalin MG, Katabi N, Persson M, et al. Multi-dimensional genomic analysis of myoepithelial carcinoma identifies prevalent oncogenic gene fusions. Nature communications. 2017;8:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickson BC, Antonescu CR, Argyris PP, et al. Ectomesenchymal Chondromyxoid Tumor: A Neoplasm Characterized by Recurrent RREB1-MKL2 Fusions. The American journal of surgical pathology. 2018;42:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laco J, Mottl R, Hobling W, et al. Cyclin D1 Expression in Ectomesenchymal Chondromyxoid Tumor of the Anterior Tongue. International journal of surgical pathology. 2016;24:586–594. [DOI] [PubMed] [Google Scholar]
- 20.Doyle LA, Vivero M, Fletcher CD, et al. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2014;27:390–395. [DOI] [PubMed] [Google Scholar]
- 21.Gerry D, Fox NF, Spruill LS, et al. Liposarcoma of the head and neck: analysis of 318 cases with comparison to non-head and neck sites. Head & neck. 2014;36:393–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.