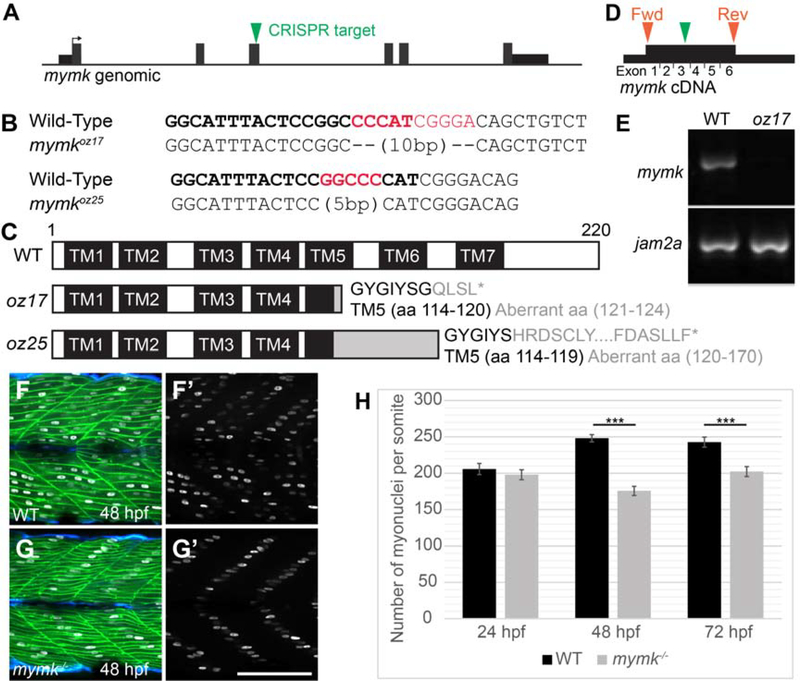
Figure 2: Embryonic skeletal muscle fusion requires mymk.
(A) The mymkCRISPR target site is within exon 3. (B) DNA lesions for mymkoz17 and mymkoz25 induced at the CRISPR target sequence (bold text), with the corresponding wild-type (WT) sequences (red text is deleted in mutant). (C) The resulting frame-shifting mutations are predicted to introduce 4 and 30 aberrant amino acids respectively (grey text), followed by a premature stop codon (*) that would truncate the protein in transmembrane (TM) domain 5 and eliminate C-terminal fusogenic elements (Millay et al. 2016). (D) Forward (fwd) and reverse (rev) primers (orange arrowheads) for RT-PCR amplify exons 1 through 6 of the cDNA including the mymkoz17 lesion site in exon 3 (green arrowhead). (E) RT-PCR of mymk and jam2a transcripts in wild-type (WT) and mymkoz17 mutant individuals at 48 hpf. (F-G’) Wild-type embryos (F, F’) contain multinucleated fast fibers at 48 hpf, while fast fibers in mymkoz17 mutant embryos (G, G’) are mononucleate, with the single nucleus in each fiber located about midway between fiber tips. myog:H2B-mRFP (white) marks myonuclei, mylfpa:lyn-cyan (green) labels fast muscle cells, and smyhc1:EGFP (aqua) labels slow muscle cells. (H) The total number of myonuclei per somite is reduced in mymk mutants compared to WT siblings beginning at 48 hpf. Myonuclei in somites 12 and 15 of each embryo were counted and averaged to get the number per somite (n= 6 for 24 hpf, n=8 for 48 hpf and 72 hpf). (Student’s t-test, p*** < 0.001). Scale bar in E’ (for D-E’) is 100 μm.