Fig. 4.
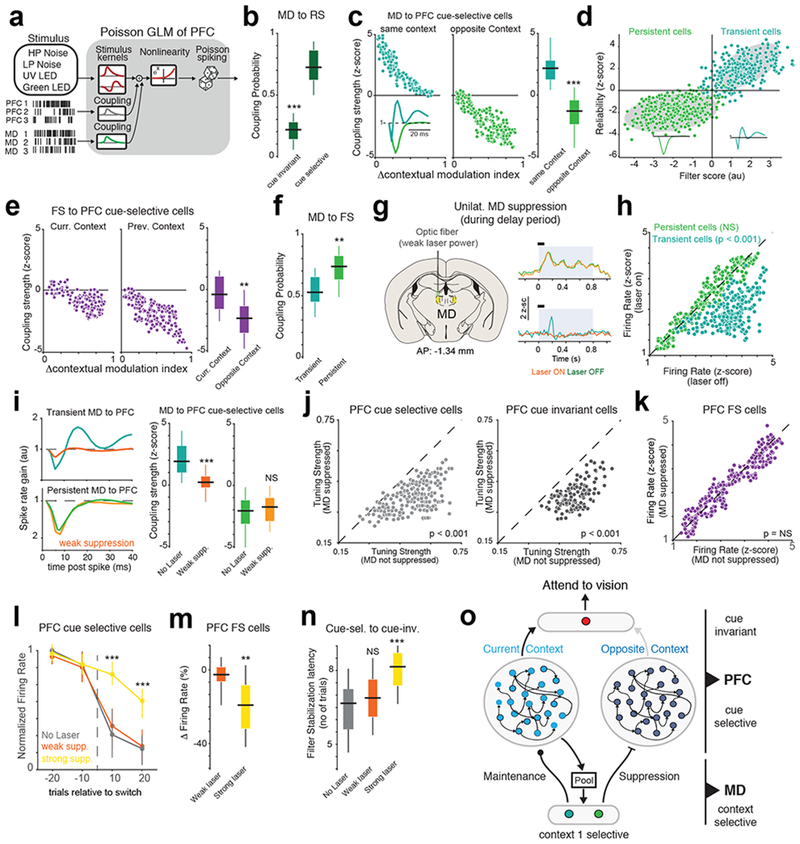
Distinct MD neurons augment and suppress context-relevant PFC representations.
(a) Schematic of Poisson GLM used to model PFC neurons including MD interactions. (b) Box-plot comparing the coupling probability between MD and PFC cue-selective (n = 230 neurons from 5 mice)) or cue-invariant neurons (n = 86 neurons from 5 mice). ***p = 0.58 x 10−5 two-way rank-sum test relative to cue-selective neurons. (c) Left: Scatter plot relating the coupling strength between MD and PFC cue-selective cell with the difference in contextual selectivity for both cells that prefer the current context or the previous context. Inset shows the two different coupling filters between MD and PFC cells. Each data point is one PFC cell from 5 mice. Right: Box-plot comparing the difference in coupling strength between MD and PFC neurons preferring the same context (n = 141 MD neurons from 5 mice) or the opposite context (n = 211 MD neurons from 5 mice). ***p = 0.89 x 10−4, two-way rank-sum test. (d) Clustering analysis relating MD-PFC coupling strength with MD spiking reliability. Each data point is one MD cell (n = 352 neurons from 5 mice). Shaded gray area is the 95% confidence interval ellipse of a Gaussian mixture model. Inset, median filter shape from each cluster. (e) Right: Scatter plot relating FS to PFC cue-selective coupling strength with difference in contextual modulation. Each data point is one PFC cell from 5 mice. Left: Box-plot comparing the difference in coupling strength between FS and PFC neurons preferring the same context (n = 141 neurons from 5 mice) or the opposite context (n = 211 neurons from 5 mice). **p = 0.31 x 10−2, two-way rank-sum test. (f) Box-plot of coupling probabilities between MD and PFC FS cells. **p = 0.78 x 10−2, two-way rank-sum test relative to transient MD neurons (n = 410 PFC FS neurons, 5 mice). (g) Left: Method for unilaterally suppressing the MD. Right: PSTHs of two example MD neurons. Persistent MD cells (top) are less affected by weak MD suppression than transient MD cells (bottom). Shaded blue area marks the duration of the laser, while the black bar marks the cueing period. (h) Scatter plot comparing the effect of weak MD suppression on the firing rates of transient (n = 260 neurons from 3 mice, p = 0.89 x 10−3) and persistent MD cells (n = 247 neurons from 3 mice, non-significant, p = 0.22, Friedman test between laser on and laser off trials. (i) Left: Example MD-PFC coupling filters with (orange) and without (green) MD suppression. Right: Box-plot comparing the effect of MD suppression on the coupling strength between transient and persistent MD cells and PFC cue selective neurons (n = 177 neurons, 3 mice). ***p = 1.02 x 10−4, NS = p = 0.12, two-way rank-sum text. (j) Scatter plot showing the change in tuning strength of cue selective (left, n = 177 neurons) and cue-invariant (right, n = 127 neurons, 3 mice) neurons caused by weak MD suppression. Friedman test between laser on and laser off trials. (k) As (j) but showing no significant effect of weak MD suppression on PFC FS neurons (264 neurons, 3 mice, p = 0.81). (l) Time course of the change in normalized maximum firing rate relative to switch of PFC cells selective for cues in the first cue set. Colors indicate various levels of MD suppression. Data shown as mean +/− SEM. N = 3 biologically-independent mice. ***p < 0.0001 one-way rank-sum test relative to no laser condition. (m) Box-plot comparing the change in firing rate of PFC FS neurons with weak and strong MD suppression (n = 264 and 212 neurons from 3 mice respectively). **p = 0.71 x 10−2 one-way Kruskal-Wallis ANOVA. (n) Box-plot comparing cue-selective to cue-invariant filter stabilization latency. No laser, n = 33 sessions, Weak laser, n = 31 sessions, Strong laser = 18 sessions from 3 mice). ***p = 0.063 x 10−4, one-way Kruskal-Wallis ANOVA with post-hoc rank-sum test. (o) Cartoon summarizing the distinct effect that MD transient and persistent cells exert on the PFC. All box plots: median (line), box edges, 95% confidence interval, whiskers, range.
