Figure 2.
Five Distinct Transcriptomic Clusters Are Defined by Single-Cell RNA Sequencing of Highly Enriched AGM HSCs
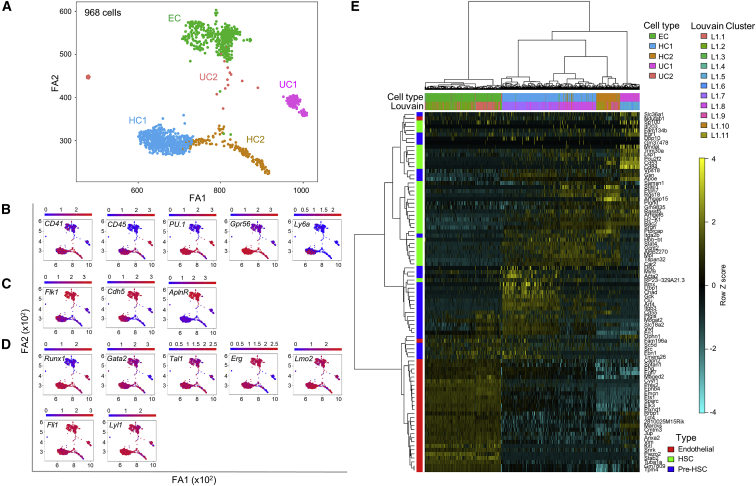
(A) SPRING visualization of merged Louvain clusters corresponding to 968 single CD31+cKithiG2med IAHC cells (n = 6 at E11, 27.6 G2V embryos, 41–49 somite pairs; n = 1 at E10, 2.7 G2V embryos, 33–34 sp). See Figure S1B for Louvain clustering. Five clusters emerge based on the cell-lineage expression characteristics and hierarchical clustering distances. EC, endothelial-like cluster; HC1, hematopoietic cluster 1; HC2, hematopoietic cluster 2; UC1, unknown cluster 1; UC2, unknown cluster 2.
(B–D) Cells were colored according to the expression of (B) hematopoietic markers, (C) endothelial markers, and (D) hematopoietic "heptad" transcription factors.
(E) Heatmap and hierarchical clustering of the dataset of 968 CD31+cKithiG2med index-sorted cells using endothelial-, pre-HSC-, and HSC-specific genes (indicated on the left in red, blue, and green, respectively) obtained from a published dataset of phenotypically enriched endothelial cells, pre-HSCs (T1+T2), and E12/E14 HSCs (Zhou et al., 2016). Louvain identity (L1.1 to L1.11) and merged clusters (EC, HC1, HC2, UC1, and UC2) are also indicated (top).