FIGURE 2.
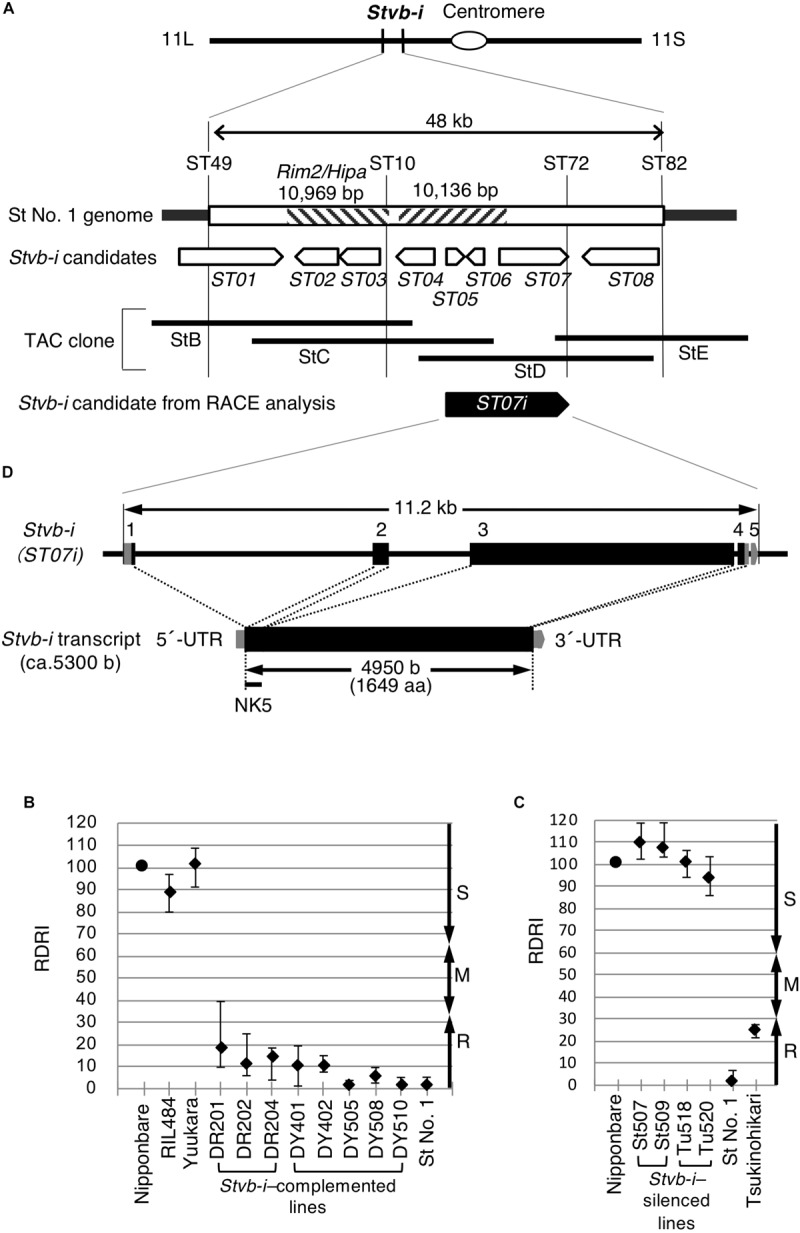
Map-based cloning of the Stvb-i gene. (A) Fine physical map of the 48-kb Stvb-i region, structure of the Stvb-i locus, and the Stvb-i candidate genes. Pentagons represent the predicted candidate genes and the identified candidate gene, ST07i, and their direction. A Rim2/Hipa transposon is present in St No. 1 but not in Nipponbare. In the Nipponbare genome, a 10,136-bp segment is substituted by a 6,383-bp segment with low sequence identity. (B) Average disease rating indexes of StD lines relative to that of Nipponbare (RDRI) (n = 6; capped bars show the ranges). Control cultivars were Nipponbare (susceptible) and St No. 1 (resistant). (C) Average RDRIs of RNAi lines (n = 3) compared with Nipponbare and St No. 1. (D) Structure of Stvb-i. Coding regions are shown in black and the untranscribed regions (UTRs) in grays. NK5 represents a trigger fragment for the RNAi construct.
