Figure 6.
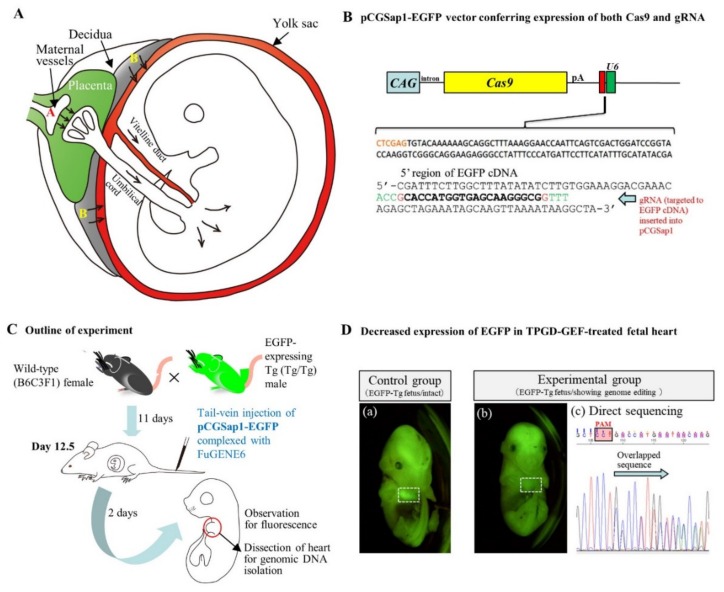
Transplacental gene delivery (TPGD) on day 12.5 (day 0 of pregnancy is defined as the day a vaginal plug is found): (A) Hypothetical mechanism of TPGD suggested by Kikuchi et al. [89] and Nakamura et al. [88]. When the TPGD is performed at day 12.5 (the time when placental circulation is established), the intravenously injected plasmid DNA/lipid complexes may be transferred via at least two routes from maternal blood to the fetus. One route is via placenta to the embryo, as indicated by the arrows (area A). Injected plasmid DNA is transferred beyond the blood-placenta barrier and enters the umbilical cord. The other route is from the decidua to the yolk sac, as depicted by the arrows (area B). Some DNA become trapped in the yolk sac and is transferred to the embryo via the vitelline circulation. (B) Structure of pCGSap1-EGFP, an all-in-one type plasmid, capable of expressing both Cas9 and gRNA (targeted to EGFP cDNA) simultaneously: This plasmid has been used for genome editing of EGFP-expressing fetal cardiac cells in TPGD-GEF (cited from Nakamura et al. [88] under permission of MDPI, publisher of the International Journal of Molecular Science). (C) Schematic representation of the experimental outline of TPGD-GEF. At day 12.5 of pregnancy, a solution containing plasmid DNA complexed with gene delivery reagent (i.e., FuGENE6) was intravenously administered to the pregnant female mice. Two days after in vivo transfection, fetuses were dissected to check the presence of the introduced DNA and its expression. (D) Decreased expression of EGFP-derived fluorescence in the heart of a TPGD-GEF-treated fetus. (a) Intact control fetus. The heart (enclosed by white dashed box) exhibited strong fluorescence. (b) TPGD-GEF-treated fetus exhibiting reduced fluorescence in its heart (enclosed by white dashed box). (c) Sequence analysis of PCR products (corresponding to the 5′ region of the EGFP sequence) from the fetus (b). Overlapping electrophoretograms (indicated by arrows) immediately upstream of the protospacer adjacent motif (PAM) indicate mosaic pattern of mutations.