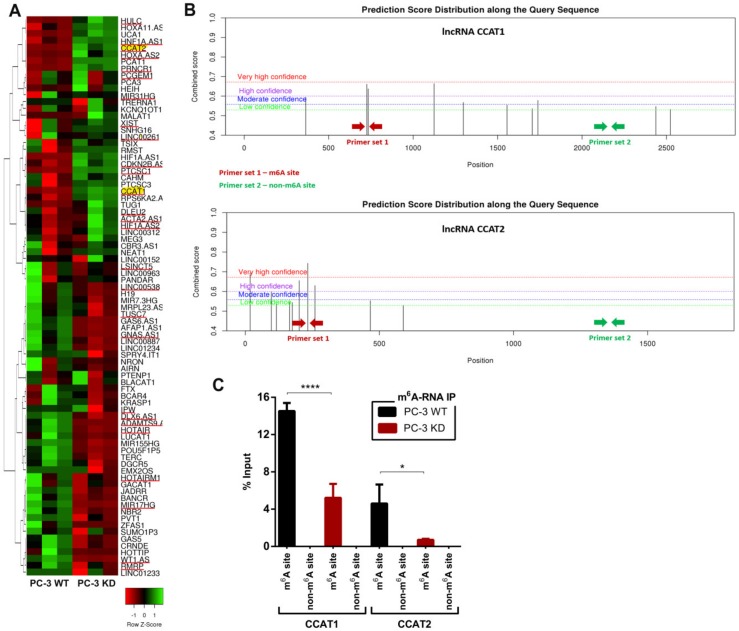
Figure 5.
VIRMA expression endorses long non-coding RNAs (lncRNAs) expression in m6A-dependent way. (A) Heat map and clustering analysis of lncRNAs expression in PC-3WT and PC-3KD cells, 3 biological replicates per condition. The 27 lncRNAs differentially expressed are underline red and the selected ones are further highlighted in yellow; (B) Potential m6A binding sites at lncRNAs CCAT1 and CCAT2 predicted with high and very high confidence (red and purple lines) by SRAMP (http://www.cuilab.cn/sramp/) [19]. Red and green arrows indicate the regions corresponding to PCR amplicons used for lncRNA quantification in immunoprecipitation experiments; (C) Abundance CCAT1 and CCAT2 transcripts in m6A-RNA IP pools from PC-3WT and PC-3KD cells. After purification precipitated RNA was used as a template for reverse transcription and real-time PCR with primers as indicated in 5b. Data is represented as percentage of input; **** p < 0.0001 and * p < 0.05, Mann–Whitney U test.