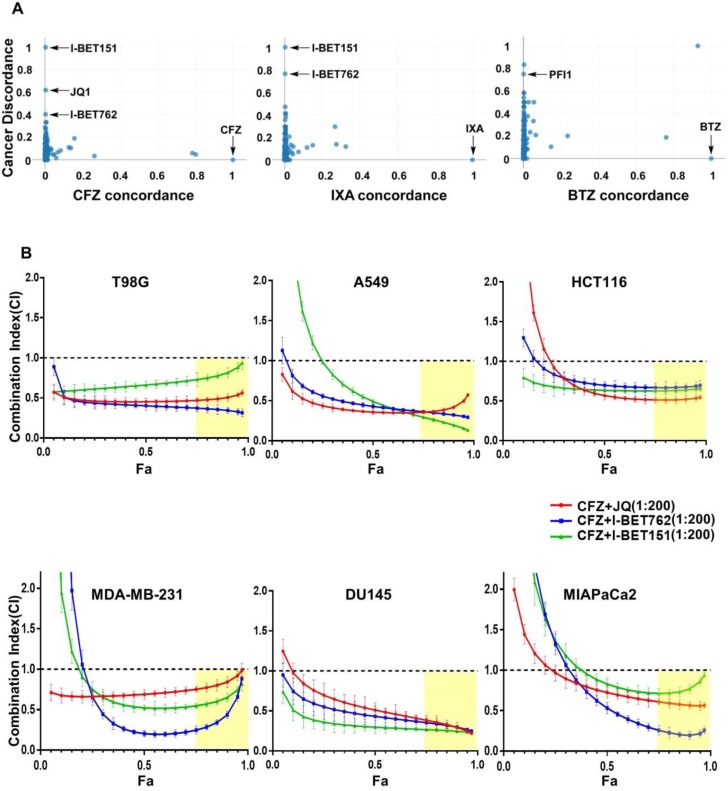
Figure 1.
Synergistic interaction between proteasome and BET inhibitors in various cancer cells. (A) SynergySeq online platform was used to identify potential drugs that can synergize with proteasome inhibitors in cancer. “cancer discordance”, a measure of the ability of a drug to reverse cancer gene expression signature to a normal state, is shown on the y-axis. The level of similarity of a drug to the reference proteasome inhibitor drugs carfilzomib (CFZ), ixazomib-citrate (IXA), and bortezomib (BTZ) is shown as “concordance” values on the x-axis; (B) T98G, A549, HCT116, MDA-MB-231, DU145, and MIAPaCa2 cells were treated with different doses of CFZ (0.5, 2, 8, and 32 nM), along with one of the BET inhibitors (I-BET762, I-BET151, and JQ1) in different doses (0.1, 0.4, 1.6, and 6.4 μM) as indicated for 72 h. In these combination treatments, the ratio of CFZ to BET inhibitors was maintained at 1:200. The combination index (CI) and fraction affected (Fa) values were determined using CompuSyn software from cell viability data and are shown in these plots. The results are shown as mean ± SD, n = 3. CI < 1.0 indicates synergism, CI = 1.0 indicates additive effect, and CI > 1.0 indicates antagonism. The regions highlighted in yellow are synergistic (CI < 1.0) at optimal Fa > 0.75.