FIGURE 2.
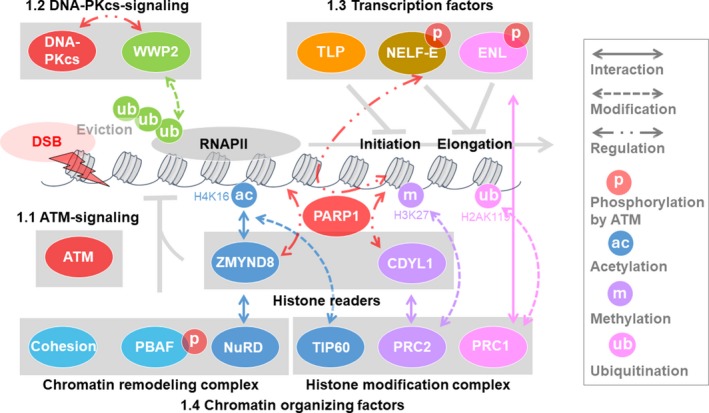
Influence of double‐strand breaks (DSBs) on transcription. DSB‐induced transcriptional repression prevents genome rearrangement and tumorigenesis. (1.1) Ataxia telangiectasia mutated (ATM) and (1.2) DNA‐dependent protein kinase catalytic subunit (DNA‐PKcs) regulate DSB‐induced transcriptional repression. (1.3) Transcription factors also regulate DSB‐induced transcriptional repression. TBP‐like protein (TLP) represses transcription globally upon transcriptional initiation. Negative elongation factor E (NELF‐E) is recruited to RNA polymerase II (RNAPII) in a poly [ADP‐ribose] polymerase 1 (PARP1)‐dependent manner and represses transcription. Eleven nineteen leukemia (ENL) in the super elongation complex recruits polycomb repressive complex 1 (PRC1) of the polycomb complex at transcriptional elongation sites in an ATM‐dependent manner and promotes histone H2A K119/120 ubiquitination and repression. (1.4) Chromatin organizing factors such as histone modification complex, chromatin remodeling complex, and cohesion are also involved in DSB‐induced transcriptional repression. Zinc finger MYND‐type containing 8 (ZMYND8) and the nucleosome remodeling and deacetylase (NuRD) complex are recruited at transcription sites via TIP60‐mediated acetylation of histones to repress transcription. Chromodomain protein, Y chromosome‐like (CDYL1) binds DSBs via H3 K9 methylation and recruits PRC2 at transcription sites to promote H3 K27 methylation for transcriptional repression. Cohesin and polybromo‐associated BRG‐/BRM‐associated factor (PBAF) repress transcription and prevent mis‐rejoining of broken DNA ends to maintain genome stability
