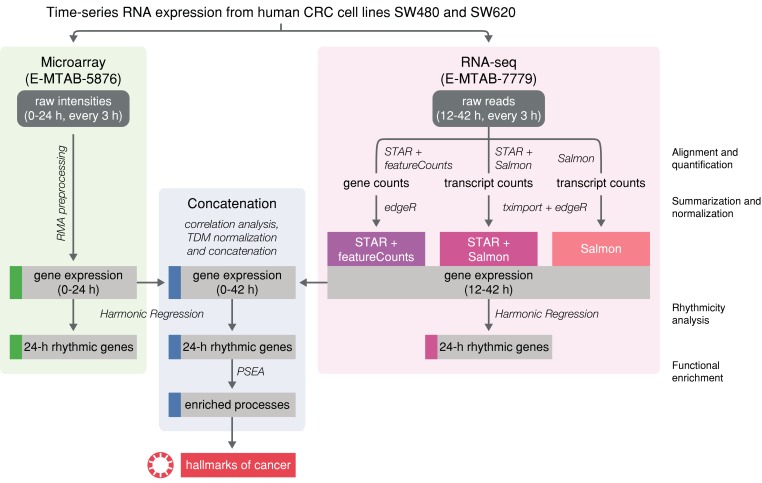
Figure 1.
Workflow for the comparison of time-series microarray and RNA-seq data from human colorectal cancer (CRC) cell lines. Two circadian datasets (microarray and RNA-seq) for the cell lines SW480 and SW620 were pre-processed to determine gene expression values. For the RNA-seq data, three different methods (STAR + featureCounts, STAR + Salmon, Salmon) for the alignment and quantification of reads were applied to determine gene expression values. A correlation analysis for shared time points revealed which RNA-seq pre-processing method yields expression values that correlate best with the microarray expression. The best-correlated RNA-seq expression dataset was then TDM-transformed to the range of the microarray data and both datasets were concatenated to gain a longer time-series. 24 h rhythmic gene sets were determined based on the three different sets of data (microarray, RNA-seq and concatenated). Biological pathways enriched for the 24 h rhythmic genes of the concatenated dataset were determined and investigated with view to hallmarks of cancer.