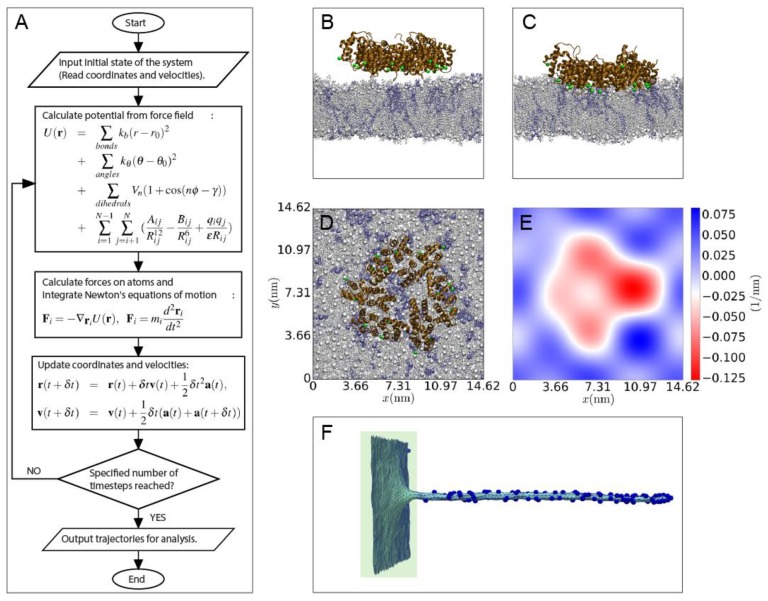
Figure 3.
(A) An overview of the molecular dynamics (MD) procedure, with a simple form of force field and velocity-verlet integration algorithm. The integration timestep in all-atom MD is usually 2 fs. (B) Initial simulation setup of the ANXA4 trimer near a POPC:POPS (4:1) bilayer. (C) Final snapshot showing the indentation of the membrane. (D) Top view of the final snapshot. (E) The 2D curvature profile for a surface passing through the center of the membrane in panel C. (F) Monte-Carlo simulation snapshot of ANXA4 protein affinity for a membrane patch (flat) and a highly curved nanotube generated by pulling a vertex from a flat membrane. Note that the proteins are depicted on the outer surface for clarity.