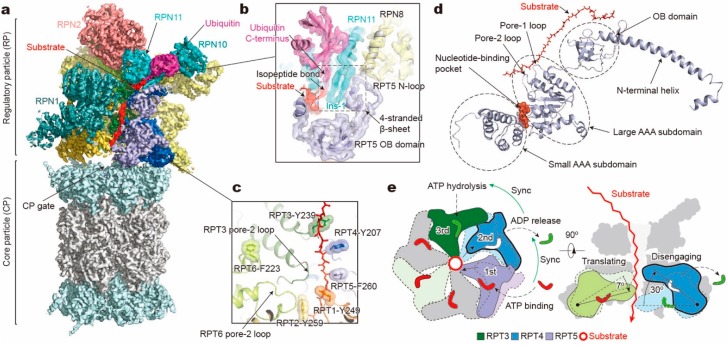
Figure 2.
Structure and translocation mechanism of the human 26S proteasome. (a) Cryo-EM structure of the substrate-bound human proteasome in state EB at 3.3 Å (EMDB ID: 9218; PDB ID: 6MSE). The RPT1 density is omitted to show the substrate density inside the ATPase ring. The RPN13 density is not observed in this structure. (b) A close-up view of the quaternary interface around the isopeptide bond between substrate and ubiquitin. (c) Architecture of pore loop staircase interacting with the substrate. Aromatic residues in pore-1 loops are labelled. (d) Molecular model of RPT5 in state EB, with ATP bound and substrate engaged. (e) Schematic of mechanical substrate translocation of proteasomal ATPases. Synchronization of nucleotide processing in three adjacent ATPases (left) causes differential vertical rigid-body rotations in each substrate-engaged ATPase that cooperatively transfer the substrate (right).