Figure 5.
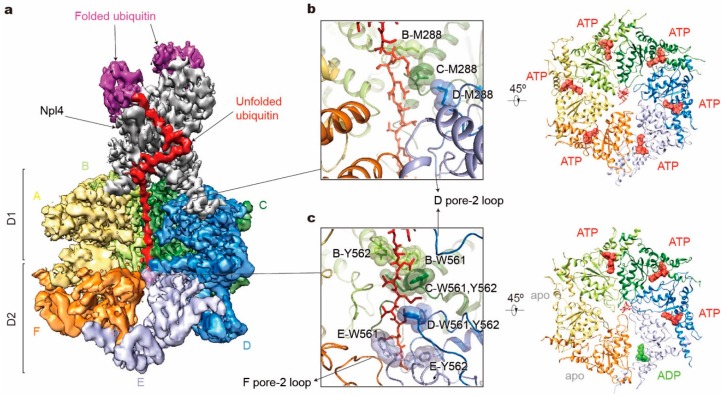
Structure illustration of the tandem hexameric ATPases of Cdc48 processing a substrate. (a) Cryo-EM density map of the Cdc48– Ufd1-Npl4 (UN)–substrate complex in the presence of ATP and with Cdc48 carrying a Walker B mutation (EMDB ID: 0665; PDB ID: 6OA9). D1 domains of subunits E and F are both omitted for clarity. Densities of Ufd1 and N domains are too vague to be shown. (b,c) Substrate interactions with the Cdc48 D1 pore (b left) and D2 pore (c left), and corresponding nucleotide states of six ATPases (right). Only key residues in pore-1 loops are labeled here.