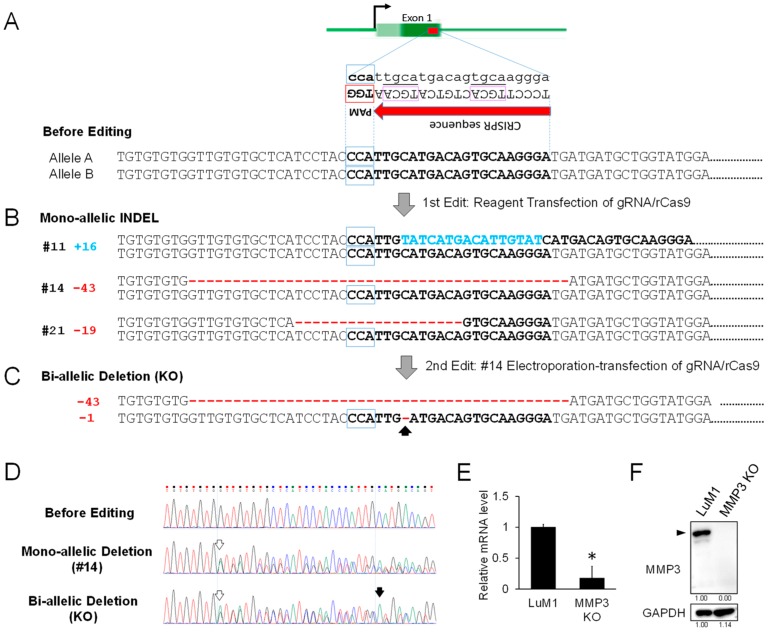
Figure 3.
Establishment of MMP3-KO cells using CRISPR/Cas9 genome editing technology. (A) Schemes representing the targeting of exon 1 in the Mmp3 gene. Top, a scheme of Mmp3 gene. Dark green box, a coding region of exon 1 including the target sequence. Bright green box, 5′-untranslated region. Middle, a CRISPR sequence containing 5′-TGCA-3′ repeat enclosed with purple square, tailed with PAM sequence (TGG) in the antisense strand. Bottom, a partial sequence of Mmp3 exon 1 including the target sequence (shown in bold). (B) The sequences of mono-allelic INDEL clones. Three types of mono-allelic INDEL clones were obtained. Clone #11 contained a 16-bp insertion sequence, shown in blue. Clones #14 and #21 conferred 43-bp and 19-bp deletions, respectively, shown in red. (C) DNA sequences of the bi-allelic deletion clone. This clone was obtained from the clone #14 with an additional single nucleotide deletion in the counterpart allele, as indicated by an arrow. (D) DNA sequences before editing, of mono-allelic deletion (#14 clone) and bi-allelic deletion (KO). White arrows, the position of the first deletion. Black arrow, the position of the single nucleotide deletion in the 2nd edit. (E) RT-qPCR analysis of Mmp3 mRNA in the parental LuM1 cells and the MMP3-KO cells. The mRNA level of Mmp3 was decreased while the mutant mRNA of Mmp3 was detectable. The relative mRNA levels were normalized to Gapdh, an internal control. * p < 0.05, n = 6. (F) Western blot showing MMP3 in the cell lysates. Arrowhead indicates full-length MMP3 (54 kD). The experiments were repeated thrice in Figure 3E,F.