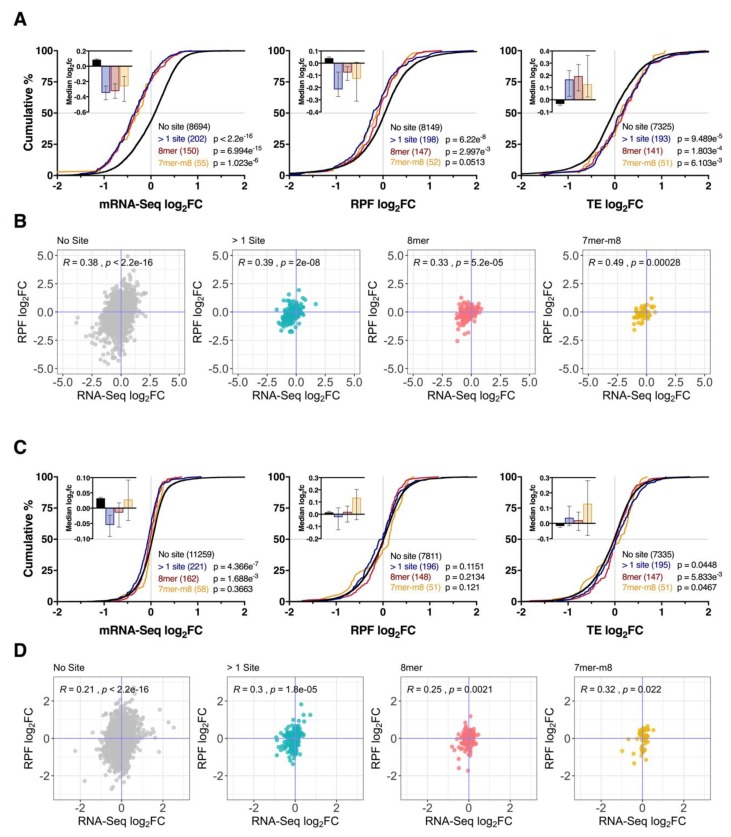
Figure 5.
Coding sequence binding sites function similarly to 3′UTR sites. Potential miR-1271-5p binding sites housed within mRNA coding sequences were predicted using custom prediction scripts provided by TargetScan with interactions exhibiting a total context+ score < −0.2 retained for further analysis. (A) Cumulative distribution analysis of mRNA, RPF, and TE expression for target genes with predicted CDS sites after miR-1271-5p overexpression. Note that, for this analysis, no 7mer-1a interactions passed context score filtration. Groups were compared to genes with no site via two-sided Kolmogorov-Smirnov test. Median log2 fold-changes ±95% CI reported top left. (B) Scatter plots comparing mRNA and RPF log2 fold changes for genes analysed in the panel (A) with Pearson’s correlation coefficients and p values reported in the top left. (C,D) As in (A,B) except after miR-1271-5p knockdown.