Abstract
Studies in the Amazon are being intensified to evaluate the alterations in the microbial communities of soils and sediments in the face of increasing deforestation and land-use changes in the region. However, since these environments present highly heterogeneous physicochemical properties, including contaminants that hinder nucleic acids isolation and downstream techniques, the development of best molecular practices is crucial. This work aimed to optimize standard protocols for DNA extraction and gene quantification by quantitative real-time PCR (qPCR) based on natural and anthropogenic soils and sediments (primary forest, pasture, Amazonian Dark Earth, and várzea, a seasonally flooded area) of the Eastern Amazon. Our modified extraction protocol increased the fluorometric DNA concentration by 48%, reaching twice the original amount for most of the pasture and várzea samples, and the 260/280 purity ratio by 15% to values between 1.8 to 2.0, considered ideal for DNA. The addition of bovine serum albumin in the qPCR reaction improved the quantification of the 16S rRNA genes of Archaea and Bacteria and its precision among technical replicates, as well as allowed their detection in previously non-amplifiable samples. It is concluded that the changes made in the protocols improved the parameters of the DNA samples and their amplification, thus increasing the reliability of microbial communities’ analysis and its ecological interpretations.
Keywords: Microbial ecology, Soil science, Soil biology, Biodiversity, Microbiology, Bacteria, Microbial genomics, Microorganism, Polymerase chain reaction, Molecular biology, Amazonian soils, DNA extraction, Quantitative real-time PCR., Archaeal communities, Bacterial communities
Microbial Ecology; Soil Science; Soil Biology; Biodiversity; Microbiology; Bacteria; Microbial Genomics; Microorganism; Polymerase Chain Reaction; Molecular Biology; Amazonian soils; DNA extraction; Quantitative real-time PCR.; Archaeal communities; bacterial communities.
1. Introduction
Studies on the microbial communities in Amazonian soils and sediments have been intensified to understand the impacts of deforestation and land-use change on their taxonomic and functional diversity (e.g., Rodrigues et al., 2013; Paula et al., 2014; Lammel et al., 2015; Mendes et al., 2015; Navarrete et al., 2015a; Meyer et al., 2017). However, these environments are complex in their physicochemical properties and contain potential organic and inorganic contaminants to nucleic acids isolation that cannot be completely removed by most extraction methods, remaining in the DNA samples and hindering downstream techniques (Moreira, 1998). Thus, the development of best practices is imperative to overcome these issues in molecular studies.
The extraction of DNA from environmental samples can be performed using direct or indirect (which involves an initial cell extraction step) approaches (Gabor et al., 2003) and requires lysis through physical, chemical, and/or enzymatic methods to disrupt the cell walls and membranes of the microorganisms and release their nucleic acids into the medium; followed by the removal of cell fragments, DNA capture and purification from contaminants (Roose-Amsaleg et al., 2001). Numerous studies have compared the most important extraction methods for soils and sediments (e.g., Stach et al., 2001; Carrigg et al., 2007; İnceoǧlu et al., 2010; Terrat et al., 2012; Leite et al., 2014; Devi et al., 2015), aiming to obtain DNA samples of high concentration and purity and consequently generate low-biased representations of their microbial communities (Robe et al., 2003). These include commercial kits, listed by Dhaliwal (2013), which are often used due to their practicality and optimized features for several environments.
Following extraction, several contaminants such as clay minerals, debris, proteins, humic substances, phenolic compounds, salts, and metal ions can still be present in environmental DNA samples (Wilson, 1997; Griffiths et al., 2000; Schrader et al., 2012; Leite et al., 2014; Narayan et al., 2016). Thus, an additional purification step is often required but can be expensive, time-consuming, and lead to DNA loss (Kreader, 1996). Alternatively, additives can be used in PCR-based methods to relieve their inhibition, including in quantitative real-time PCR (qPCR) reactions (Schriewer et al., 2011; Albers et al., 2013; Sidstedt et al., 2015). The most used PCR additive is the bovine serum albumin (BSA), a transport protein that can bind to lipids and organic molecules, thus being able to reduce several types of inhibition (Hedman and Rådström, 2013).
Up to date, there are no comprehensive DNA protocols optimized for Amazonian soils. Therefore, this work aimed to improve standard protocols for DNA extraction and qPCR based on natural and anthropogenic soils and sediments (primary forest, pasture, Amazonian Dark Earth, and várzea, a seasonally flooded area) of the Eastern Amazon through minor laboratory modifications, aiming to increase the concentration and quality of the extracted DNA and reduce the inhibition of the qPCR reactions using BSA.
2. Material and methods
2.1. Soil sampling and characterization
The sampling sites are located at the Tapajós National Forest and its surroundings, in the Pará State, Brazil, Eastern Amazon. The forest has a tropical monsoon climate (Am Köppen) and consists mostly of nutrient-poor oxisols and ultisols (Silver et al., 2000; IBAMA, 2004). This region is covered by primary, logged, and secondary forests as well as lands converted to pasture and agricultural fields, including manioc, rice, beans, corn, soybean, sugarcane, coffee, black pepper, and fruit crops (D'Antona et al., 2006; Steward, 2007). It also harbors Amazonian Dark Earths, soils resulting from human activities mainly between 2,500 to 500 BP (Neves et al., 2003) that cover about 10% of the Amazon (Mann, 2002) and are characterized by high levels of stable carbon (such as charcoal and humic substances), organic matter, and nutrients (Mann, 2002; Kämpf et al., 2003; Lehmann et al., 2003; Novotny et al., 2007; Glaser and Birk, 2012); and lowlands that periodically receive water and sediments from their adjacent rivers, which constitute 13% of the territory (Nascimento and Homma, 1984).
The sampling was conducted in May and October 2016 at four sites: (1) primary forest (PF, 3°17′44.4″S 54°57′46.7″W); (2) pasture (PA, 3°18′46.7″S 54°54′34.8″W); (3) Amazonian Dark Earth (ADE, 2°50′36.1″S 54°58′32.6″W); (4) and várzea (VA, 2°22′44.8″S 54°44′21.1″W), a type of seasonally flooded area (floodplain). At each site, three soil samples from 0 to 10 cm depth were collected. Following their transportation to the laboratory, aliquots were sent to the Department of Soil Science of the Luiz de Queiroz College of Agriculture (ESALQ/USP) for the determination of the following physicochemical properties: pH determined in 0.01 M calcium chloride (CaCl2); soil organic matter (SOM) determined by colorimetry; nitrogen (N) determined by the Kjeldahl method; phosphorus (P) extracted with ion exchange resin and determined by the colorimetric method; sulfur (S) extracted with 0.01 M calcium phosphate (Ca3(PO4)2) and determined by turbidimetry; potassium (K) extracted with ion exchange resin and determined by atomic emission spectrophotometry; calcium (Ca) and magnesium (Mg) extracted with ion exchange resin and determined by atomic absorption spectrophotometry; exchangeable aluminum (Al) extracted with 1 M potassium chloride (KCl) and determined by the colorimetric method; potential acidity (H + Al) determined with the SMP buffer; boron (B) extracted with hot water and determined by colorimetry; copper (Cu), iron (Fe), manganese (Mn), and zinc (Zn) extracted with DTPA and determined by atomic absorption spectrophotometry. Sum of bases (SB), cation-exchange capacity (CEC), base saturation (V), and aluminum saturation (m) calculations were made based on these previous results. The contents of sand, silt, and clay were determined by the hydrometer method and classified according to the United States Department of Agriculture (USDA) classification (2018).
2.2. DNA extraction and quantification
The soil DNA extraction was performed using the DNeasy PowerLyzer PowerSoil Kit (Qiagen), a widely used commercial kit for soils due to its mechanical-chemical methods for cell lysis, patented inhibitor reagent to remove organic and inorganic contaminants (comprising humic acids, cell debris, and proteins), and silica membranes for DNA capture and cleaning (MO BIO Laboratories, 2016). The total DNA of each sample was extracted by two methods: the manufacturer's and an optimized protocol, in which after adding the solution C1, the samples were vortexed for 15 min at maximum speed, and centrifuged for 3 min at 10,000 x g. Besides, all the incubations of the modified protocol after adding the solutions C2 and C3 were made at minus 20 °C (-20 °C) instead of 4 °C. The DNA samples were evaluated on a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) and a Qubit fluorometer Q32857 using the dsDNA BR Assay Kit (Thermo Fisher Scientific).
2.3. Quantitative real-time PCR of the 16S rRNA genes of Archaea and Bacteria
The 16S rRNA genes were quantified by quantitative real-time PCR through the standard-curve method using the primer pairs Arch519F (Øvreås et al., 1997) and Arch915R (Stahl and Amann, 1991) for Archaea, and 926F and 1062R (De Gregoris et al., 2011) for Bacteria. For both genes, the qPCR of each DNA sample extracted with the optimized protocol was performed in a 10-μL reaction on a StepOnePlus instrument (Thermo Fisher Scientific) in triplicate for each of the tested treatments: (1) no bovine serum albumin and (2) 0.1 μL of BSA (20 mg mL−1) (Thermo Fisher Scientific), resulting in a final concentration of 200 ng μL−1. Each 10-μL reaction mixture included 5 μL of SYBR Green ROX qPCR Master Mix (Thermo Fisher Scientific), 1 μL of each primer (5 pmol), 1 μL of DNA (10 ng), and ultra-pure H2O to complete 10 μL. The amplification conditions for Archaea consisted of 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 57 °C for 30 s and 72 °C for 50 s followed by a melting curve of 95 °C for 15 s, 57 °C for 1 min and 95 °C for 15 s; and for Bacteria, 95 °C for 10 min, 40 cycles of 95 °C for 45 s, 60 °C for 15 s and 72 °C for 20 s followed by a melting curve of 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. The results were analyzed using StepOne Software v2.3 (Thermo Fisher Scientific), exported as spreadsheets, and converted into the number of gene copies per ng of DNA.
2.4. Statistical analysis
The soil physicochemical properties were analyzed by analysis of variance (ANOVA) followed by the Tukey's post-hoc test using the agricolae package 1.2–8 (de Mendiburu, 2017) in R studio 1.0.153 (RStudio Team, 2016) and also subjected to a non-metric multidimensional scaling (NMDS) based on the Gower's distance using the vegan package 2.5–1 (Oksanen et al., 2018). The NMDS plot was generated using the ggplot2 package 3.0.0 (Wickham, 2016). The results from the NanoDrop and Qubit quantification of the DNA samples and qPCR of the 16S rRNA genes of Archaea and Bacteria were aligned rank-transformed and analyzed by a two-way mixed-design ANOVA using the ARTool package 0.10.5 (Kay and Wobbrock, 2018). Post-hoc tests (Holm-adjusted) were carried out using the lsmeans package 2.27–62 (Lenth, 2016).
3. Results and discussion
3.1. Soil physicochemical properties
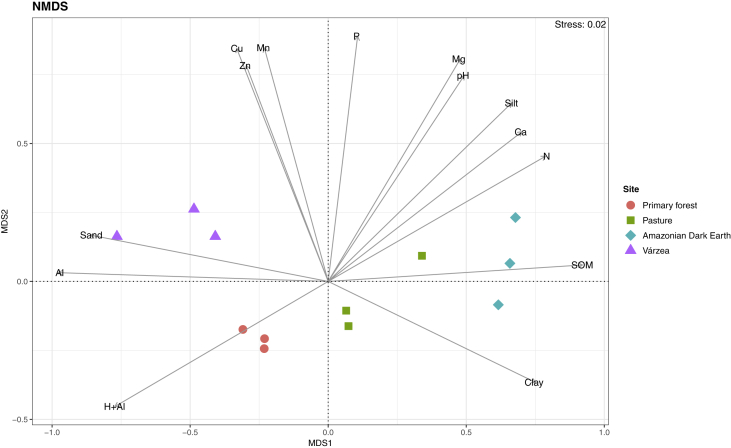
The chosen sites, which represent different Amazonian environments in this study, exhibited contrasting physicochemical properties. The sampled soils and sediments were classified as sandy clay loam (PF), clay (PA and ADE), and sandy loam (VA), according to the USDA textural classification (2018). The pH of all samples was found to be acidic, ranging from 3.5 to 5.1 (Figure 1 and Table 1). Using ANOVA followed by Tukey's post-hoc test at 0.05 level of significance, the PF site had the lowest pH and, together with VA, the highest values of Al and m. In opposition to most of the Amazonian soils, considered weathered, highly acidic, and low-fertile, ADEs are known to present large amounts of charcoal and humic substances as well as more organic matter and nutrients than their surroundings, including N, P, Ca, Mg, S, Mn, and Cu (Mann, 2002; Kämpf et al., 2003; Lehmann et al., 2003; Novotny et al., 2007; Glaser and Birk, 2012). In our study, this site presented elevated macronutrient levels, with the highest values of Ca, Mg, SB, CEC, and V among the studied soils. ADE also had higher contents of SOM and N than PF and VA, and P compared to PF. The VA site, which receives sediments from both Tapajós and Amazon rivers in the rainy season, showed the highest levels of Cu and Mn. This site also showed higher contents of K and Zn than PF and ADE.
Figure 1.
Non-metric multidimensional scaling (NMDS) based on the Gower's distance of the soil physicochemical properties of the primary forest, pasture, Amazonian Dark Earth, and várzea sites. Significant soil properties (p < 0.01) are shown in the arrows. SOM, soil organic matter; H + Al, potential acidity.
Table 1.
Mean and standard deviation of the soil chemical properties of the primary forest, pasture, Amazonian Dark Earth, and várzea sites.
| Properties | Units | Primary forest | Pasture | Amazonian Dark Earth | Várzea |
|---|---|---|---|---|---|
| pH | - | 3.53 ± 0.06 c | 4.37 ± 0.29 ab | 4.83 ± 0.23 a | 4.07 ± 0.06 b |
| SOM | g dm−3 | 32.67 ± 3.51 bc | 40.33 ± 19.66 ab | 70.67 ± 13.05 a | 7.33 ± 1.15 c |
| N | mg kg−1 | 1,822.33 ± 77.42 b | 2,837.33 ± 1,277.18 ab | 3,894.33 ± 610.26 a | 1,348.67 ± 271.14 b |
| P | mg dm−3 | 6.00 ± 1.00 b | 7.67 ± 4.62 ab | 15.67 ± 3.51 a | 14.67 ± 4.04 ab |
| S | mg dm−3 | 6.00 ± 2.00 a | 4.67 ± 1.15 a | 14.00 ± 2.00 a | 13.00 ± 12.29 a |
| K | mmolc dm−3 | 0.37 ± 0.06 c | 1.63 ± 0.85 ab | 0.67 ± 0.15 bc | 1.97 ± 0.06 a |
| Ca | mmolc dm−3 | 3.00 ± 0.00 b | 16.00 ± 13.86 b | 130.67 ± 6.66 a | 5.67 ± 1.53 b |
| Mg | mmolc dm−3 | 1.67 ± 0.58 b | 6.00 ± 4.36 b | 18.00 ± 3.00 a | 5.33 ± 1.53 b |
| Al | mmolc dm−3 | 12.33 ± 0.58 a | 4.67 ± 3.21 b | 1.00 ± 0.00 b | 16.00 ± 1.73 a |
| H + Al | mmolc dm−3 | 82.00 ± 5.20 a | 56.00 ± 3.46 ab | 33.33 ± 11.72 b | 75.33 ± 19.63 a |
| SB | mmolc dm−3 | 4.70 ± 1.21 b | 23.63 ± 17.90 b | 149.33 ± 3.64 a | 12.97 ± 3.05 b |
| CEC | mmolc dm−3 | 86.70 ± 3.98 b | 79.63 ± 14.44 b | 182.67 ± 8.09 a | 88.30 ± 22.31 b |
| V | % | 5.33 ± 1.15 c | 27.67 ± 15.89 b | 82.00 ± 5.29 a | 14.67 ± 2.08 bc |
| m | % | 72.67 ± 6.35 a | 22.67 ± 18.01 b | 1.00 ± 0.00 b | 55.67 ± 3.79 a |
| B | mg dm−3 | 0.41 ± 0.03 a | 0.41 ± 0.04 a | 0.22 ± 0.08 b | 0.23 ± 0.02 b |
| Cu | mg dm−3 | 0.23 ± 0.06 b | 0.53 ± 0.32 b | 0.37 ± 0.21 b | 3.53 ± 1.38 a |
| Fe | mg dm−3 | 99.67 ± 43.68 a | 87.67 ± 17.01 a | 67.00 ± 31.19 a | 118.67 ± 9.29 a |
| Mn | mg dm−3 | 3.53 ± 2.92 b | 5.47 ± 6.62 b | 15.93 ± 8.24 b | 115.10 ± 62.73 a |
| Zn | mg dm−3 | 0.47 ± 0.15 b | 1.33 ± 1.29 ab | 0.23 ± 0.06 b | 3.03 ± 1.19 a |
Different letters indicate significant differences among sites according to the Tukey's post-hoc test (p < 0.05). SOM, soil organic matter; H + Al, potential acidity; SB, sum of bases; CEC, cation-exchange capacity; V, base saturation; m, aluminum saturation.
The soil environment results from multiple interacting factors, including texture, pH, and nutrient content (Robe et al., 2003). A number of studies have revealed the importance of physicochemical properties on the structure and functioning of tropical soil microbial communities (Lammel et al., 2018; Merloti et al., 2019; Pedrinho et al., 2019), although access to this diversity can be challenging as soil characteristics (including clay and organic matter contents) influence the efficiency of microbial DNA recovery (Roose-Amsaleg et al., 2001). In this sense, the results of the physicochemical analysis demonstrated that even nearby areas within the Amazon biome provide different environments for soil microbial communities; and therefore, it is imperative to establish a unique DNA extraction protocol that takes into account all these heterogeneous properties.
3.2. DNA concentration and purity
The choice of the DNA extraction method is a crucial step in molecular studies, which can affect the detection of microbial communities’ structure and composition (İnceoǧlu et al., 2010; Terrat et al., 2012; Hallmaier-Wacker et al., 2018). The DNeasy PowerLyzer PowerSoil Kit (Qiagen) is one of the most used commercial kits for direct DNA extraction from tropical soils (e.g., Navarrete et al., 2015b; de Araujo et al., 2017; Goss-Souza et al., 2017; Valadares-Pereira et al., 2017; Lammel et al., 2018; Portilho et al., 2018), including Amazonian samples (e.g., Mirza et al., 2014; Paula et al., 2014; Hamaoui et al., 2016; Meyer et al., 2017). However, considering the heterogeneous physicochemical properties of these environments and that each soil type should have its own optimized DNA protocol respecting its unique composition and biomass abundance (Narayan et al., 2016), no work has been done so far to improve this extraction method for samples from several Amazonian land-uses. We choose three soils and one sediment of the Eastern Amazon to compare the protocol of the DNeasy PowerLyzer PowerSoil Kit (Qiagen) with an optimized version in which minor laboratory modifications were made regarding the duration and temperature of its steps.
The DNA concentration measured by Qubit increased (F1,8 = 10.462, p = 0.012) using the optimized protocol, reaching almost twice the original amount for most of the PA and VA samples (Table 2), also noticed as ticker bands on agarose gel electrophoresis (data not shown). This improvement was not so clearly observed in the NanoDrop data; however, spectrophotometric methods are considered less specific and accurate in comparison with the Qubit system (O'Neill et al., 2011). The results from both quantification methods varied according to the studied site (Qubit: F3,8 = 6.617, p = 0.015; NanoDrop: F3,8 = 6.161, p = 0.018), with post-hoc tests (Holm-adjusted) indicating a significant difference (p < 0.05) between ADE and VA.
Table 2.
Mean and standard deviation of the Qubit concentration (ng μL−1), NanoDrop concentration (ng μL−1), 260/280 and 260/230 ratios of the DNA samples from the primary forest, pasture, Amazonian Dark Earth, and várzea sites; followed by the results (degrees of freedom, F-values, and p-values) of the two-way mixed-design ANOVA of the aligned rank-transformed data.
| Site | Protocol | Qubit Concentration | NanoDrop Concentration | 260/280 | 260/230 |
|---|---|---|---|---|---|
| Primary forest | Original | 9.94 ± 2.37 | 15.13 ± 3.01 | 1.63 ± 0.07 | 1.55 ± 0.45 |
| Modified | 11.00 ± 1.39 | 16.87 ± 1.78 | 1.88 ± 0.09 | 1.14 ± 0.19 | |
| Pasture | Original | 10.57 ± 6.91 | 17.32 ± 14.01 | 1.69 ± 0.10 | 1.86 ± 0.17 |
| Modified | 19.40 ± 14.38 | 16.56 ± 7.34 | 1.96 ± 0.12 | 1.18 ± 0.46 | |
| Amazonian Dark Earth | Original | 33.57 ± 5.86 | 28.81 ± 3.80 | 1.77 ± 0.01 | 1.87 ± 0.05 |
| Modified | 33.77 ± 5.29 | 29.56 ± 4.95 | 1.90 ± 0.03 | 1.67 ± 0.16 | |
| Várzea | Original | 5.81 ± 1.28 | 10.12 ± 1.19 | 1.62 ± 0.05 | 1.22 ± 0.26 |
| Modified |
10.63 ± 1.87 |
9.49 ± 2.27 |
1.95 ± 0.09 |
0.99 ± 0.07 |
|
| Site | 3 | 6.617∗ | 6.161∗ | 1.180 | 7.817∗∗ |
| Protocol | 1 | 10.462∗ | 0.075 | 107.301∗∗∗ | 13.256∗∗ |
| Site × Protocol | 3 | 1.730 | 0.090 | 4.242∗ | 1.053 |
∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
The 260/280 ratio increased (F1,8 = 107.301, p < 0.001) using the modified protocol for all samples to values between 1.8 and 2.0, considered pure for DNA (Mathieson and Thomas, 2013), but it also varied according to the land-use since a significant interaction (F3,8 = 4.242, p = 0.045) between protocol and site was found. The 260/230 ratio, a secondary measure of nucleic acid purity that should usually be higher (between 2.0 and 2.2) than its respective 260/280, presented low values for both original and modified protocols, which can be related to the use of column-based extraction methods; as well as indicate the presence of contaminants, such as humic acids, urea, carbohydrates, proteins, lipids, salts, guanidine, phenol, and EDTA (Thermo Fisher Scientific, 2010; Lucena-Aguilar et al., 2016). This ratio was influenced by protocol (F1,8 = 13.256, p = 0.007) and site (F3,8 = 7.817, p = 0.009) and, similar to the concentration results, post-hoc tests (Holm-adjusted) also showed a significant difference (p < 0.05) between ADE and VA.
Soil DNA extraction is a challenging procedure since microbial cells can bind to its particles (Lindahl and Bakken, 1995) as well as DNA can be adsorbed on sands, clays, and macromolecules, such as humic acids (Nielsen et al., 2006). The adsorption of DNA is affected by several factors, including the size of its fragments (Ogram et al., 1994), content and type of clay (Cai et al., 2006), and pH (Greaves and Wilson, 1969; Khanna and Stotzky, 1992), being favored by acidic conditions, as found in our samples. However, the changes in vortexing and centrifugation times, the latter previously recommended by MO BIO Laboratories (2016) for clayey soils, allowed the greater breakdown of soil aggregates and microbial cells, release of nucleic acids as well as the better separation of soil debris. Besides, the freezer incubations after adding the solutions C2 and C3 were essential to ensure the recommended low-temperature conditions, avoiding the degradation of the DNA samples.
3.3. Quantification of the 16S rRNA genes of Archaea and Bacteria
PCR inhibitors compose a large group of organic and inorganic substances that can be found in the sample itself or introduced during its transport, processing, and nucleic acids extraction (Schrader et al., 2012), and which can attenuate the amplification of DNA in several degrees or inhibit it completely (Moreira, 1998). The presence of inhibitors is especially problematic for qPCR since this method is considered to be much more sensitive than the regular (endpoint) PCR so that even the smallest inhibition can generate unreliable results (Albers et al., 2013). In soil and sediment samples, humic acids are the most common contaminant co-extracted with DNA (Yeates et al., 1998), which contain phenolic groups that can denature biological molecules by bonding to amides or oxidize to form a quinone that covalently bonds to proteins and DNA (Young et al., 1993). In addition, humic acids can inhibit DNA polymerase and chelate magnesium ions, an essential cofactor for its activity (Tsai and Olson, 1992a, 1992b; Sidstedt et al., 2015). Different molecular mechanisms associated with their effect on SYBR Green assays have also been proposed (Zipper et al., 2003).
Humic acids are not easily removed from DNA extracts by purification methods (Lakay et al., 2007; Sagova-Mareckova et al., 2008), but the impacts of these contaminants on qPCR amplification can be relieved using BSA, a widely used additive for environmental samples that contain potential inhibitors, in concentrations ranging from 40 to 400 ng μL−1 (Schriewer et al., 2011). In our samples (all samples were previously adjusted so that each amplification reaction for both genes contained 10 ng of DNA), the BSA addition increased (F1,8 = 54.966, p < 0.001) the quantification of the 16S rRNA gene of Bacteria (Table 3). For the archaeal 16S rRNA gene, a significant effect of the treatment (F1,8 = 80.062, p < 0.001), studied site (F3,8 = 6.411, p = 0.016) and their interaction (F3,8 = 9.148, p = 0.006) was also observed. Post-hoc analysis (Holm-adjusted) showed that the difference in the archaeal abundance between the BSA treatment and control from the ADE site was significantly different (p < 0.05) compared to the differences found in PF and PA sites.
Table 3.
Mean and standard deviation of the qPCR quantification (copies ng−1 DNA) of the 16S rRNA genes of Archaea and Bacteria for the primary forest, pasture, Amazonian Dark Earth, and várzea sites using no (control) and 200 ng μL−1 of BSA; followed by the results (degrees of freedom, F-values, and p-values) of the two-way mixed-design ANOVA of the aligned rank-transformed data.
| Site | Treatment | 16S rRNA of Archaea | 16S rRNA of Bacteria |
|---|---|---|---|
| Primary forest | Control | 9.48E+02 ± 1.11E+03 | 1.44E+04 ± 2.41E+04 |
| BSA | 1.51E+04 ± 3.51E+03 | 4.98E+05 ± 1.40E+05 | |
| Pasture | Control | 3.52E+03 ± 1.58E+03 | 3.99E+04 ± 7.41E+03 |
| BSA | 2.43E+04 ± 1.01E+04 | 6.46E+05 ± 3.55E+05 | |
| Amazonian Dark Earth | Control | 5.38E+02 ± 7.77E+02 | 4.70E+03 ± 8.14E+03 |
| BSA | 4.51E+04 ± 8.84E+03 | 2.10E+05 ± 1.84E+05 | |
| Várzea | Control | 6.71E+02 ± 6.04E+02 | 1.26E+04 ± 1.09E+04 |
| BSA |
2.65E+04 ± 7.06E+03 |
2.17E+05 ± 1.58E+05 |
|
| Site | 3 | 6.411∗ | 3.187 |
| Treatment | 1 | 80.062∗∗∗ | 54.966∗∗∗ |
| Site × Treatment | 3 | 9.148∗∗ | 2.026 |
∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
Besides highly increasing gene abundance, the BSA addition allowed the detection of both genes in non-amplifiable DNA samples (without the additive) and improved the precision of the quantification among technical replicates, ensuring the replicability of the results. Although the effect of BSA on relieving the influence of inhibitors can vary according to the DNA polymerase used in the qPCR reaction (Albers et al., 2013), considering the conditions applied to this study, this additive was essential for the gene quantification from samples complex in inhibitors, such as the soils and sediments of the Amazon. Higher BSA concentrations (400 ng μL−1) were also tested and showed to decrease the quantification for most samples in comparison to our BSA treatment, but the results varied according to the site and target gene (data not shown).
4. Conclusion
We conclude that our optimized extraction protocol increased the concentration and purity of the DNA samples, as well as the BSA addition in the qPCR reaction allowed better gene amplification and quantification, thus increasing the reliability of the molecular data and the inferences to be drawn from them regarding the microbial communities in soils and sediments of the Amazon.
Declarations
Author contribution statement
Andressa Monteiro Venturini, Fernanda Mancini Nakamura, Júlia Brandão Gontijo: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Aline Giovana da França: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Caio Augusto Yoshiura: Analyzed and interpreted the data; Wrote the paper.
Jéssica Adriele Mandro: Conceived and designed the experiments; Performed the experiments.
Siu Mui Tsai: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the São Paulo Research Foundation (FAPESP grants 2014/50320-4, 2015/08564-6, 2015/12282-6, 2015/13546-7, 2015/19979-2, 2015/23758-1, and 2018/14974-0), the National Council for Scientific and Technological Development (CNPq grants 149662/2014-9, 140032/2015-0, 133769/2015-1, and 311008/2016-0), and the Coordination for the Improvement of Higher Education Personnel - Brasil (CAPES) - Finance Code 001.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Albers C.N., Jensen A., Bælum J., Jacobsen C.S. Inhibition of DNA polymerases used in q-PCR by structurally different soil-derived humic substances. Geomicrobiol. J. 2013;30:675–681. [Google Scholar]
- Cai P., Huang Q., Zhang X., Chen H. Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol. Soil Biol. Biochem. 2006;38:471–476. [Google Scholar]
- Carrigg C., Rice O., Kavanagh S., Collins G., O’Flaherty V. DNA extraction method affects microbial community profiles from soils and sediment. Appl. Microbiol. Biotechnol. 2007;77:955–964. doi: 10.1007/s00253-007-1219-y. [DOI] [PubMed] [Google Scholar]
- de Araujo A.S.F., Bezerra W.M., dos Santos V.M., Rocha S.M.B., Carvalho N.S., de Lyra M.C.C.P., Figueiredo M.V.B., Lopes A.C.A., Melo V.M.M. Distinct bacterial communities across a gradient of vegetation from a preserved Brazilian Cerrado. Antonie Leeuwenhoek. 2017;110:457–469. doi: 10.1007/s10482-016-0815-1. [DOI] [PubMed] [Google Scholar]
- De Gregoris T.B., Aldred N., Clare A.S., Burgess J.G. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods. 2011;86:351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- de Mendiburu F. 2017. Agricolae: Statistical Procedures for Agricultural Research. R Package Version 1.2-8.https://CRAN.R-project.org/package=agricolae [Google Scholar]
- Devi S.G., Fathima A.A., Radha S., Arunraj R., Curtis W.R., Ramya M. A rapid and economical method for efficient DNA extraction from diverse soils suitable for metagenomic applications. PloS One. 2015;10 doi: 10.1371/journal.pone.0132441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal A. DNA extraction and purification. Mater. Methods. 2013;3 [Google Scholar]
- D’Antona A.O., VanWey L.K., Hayashi C.M. Property size and land cover change in the Brazilian Amazon. Popul. Environ. 2006;27:373–396. [Google Scholar]
- Gabor E.M., de Vries E.J., Janssen D.B. Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol. Ecol. 2003;44:153–163. doi: 10.1016/S0168-6496(02)00462-2. [DOI] [PubMed] [Google Scholar]
- Glaser B., Birk J.J. State of the scientific knowledge on properties and genesis of Anthropogenic Dark Earths in Central Amazonia (terra preta de índio) Geochem. Cosmochim. Acta. 2012;82:39–51. [Google Scholar]
- Goss-Souza D., Mendes L.W., Borges C.D., Baretta D., Tsai S.M., Rodrigues J.L.M. Soil microbial community dynamics and assembly under long-term land use change. FEMS Microbiol. Ecol. 2017;93:fix109. doi: 10.1093/femsec/fix109. [DOI] [PubMed] [Google Scholar]
- Greaves M.P., Wilson M.J. The adsorption of nucleic acids by montmorillonite. Soil Biol. Biochem. 1969;1:317–323. [Google Scholar]
- Griffiths R.I., Whiteley A.S., O'Donnell A.G., Bailey M.J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA-and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000;66:5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmaier-Wacker L.K., Lueert S., Roos C., Knauf S. The impact of storage buffer, DNA extraction method, and polymerase on microbial analysis. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-24573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaoui G.S., Rodrigues J.L.M., Bohannan B.J.M., Tiedje J.M., Nüsslein K. Land-use change drives abundance and community structure alterations of thaumarchaeal ammonia oxidizers in tropical rainforest soils in Rondônia, Brazil. Appl. Soil Ecol. 2016;107:48–56. [Google Scholar]
- Hedman J., Rådström P. Overcoming inhibition in real-time diagnostic PCR. In: Wilks M., editor. PCR Detection of Microbial Pathogens. Vol. 943. Humana Press; Totowa: 2013. pp. 17–48. (Methods in Molecular Biology (Methods and Protocols)). [DOI] [PubMed] [Google Scholar]
- IBAMA . IBAMA; Brasília: 2004. Floresta Nacional do Tapajós: Plano de Manejo. Volume I - Informações Gerais. [Google Scholar]
- İnceoǧlu Ö., Hoogwout E.F., Hill P., van Elsas J.D. Effect of DNA extraction method on the apparent microbial diversity of soil. Appl. Environ. Microbiol. 2010;76:3378–3382. doi: 10.1128/AEM.02715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpf N., Woods W.I., Sombroek W., Kern D.C., Cunha T.J.F. Classification of Amazonian Dark Earths and other ancient anthropic soils. In: Lehmann J., Kern D.C., Glaser B., Woods W.I., editors. Amazonian Dark Earths: Origin, Properties, Management. Springer; Dordrecht: 2003. pp. 77–102. [Google Scholar]
- Kay M., Wobbrock J. 2018. ARTool: Aligned Rank Transform for Nonparametric Factorial ANOVAs. R Package Version 0.10.5.https://github.com/mjskay/ARTool [Google Scholar]
- Khanna M., Stotzky G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 1992;58:1930–1939. doi: 10.1128/aem.58.6.1930-1939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader C.A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakay F.M., Botha A., Prior B.A. Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. J. Appl. Microbiol. 2007;102:265–273. doi: 10.1111/j.1365-2672.2006.03052.x. [DOI] [PubMed] [Google Scholar]
- Lammel D.R., Nüsslein K., Tsai S.M., Cerri C.C. Land use, soil and litter chemistry drive bacterial community structures in samples of the rainforest and Cerrado (Brazilian Savannah) biomes in Southern Amazonia. Eur. J. Soil Biol. 2015;66:32–39. [Google Scholar]
- Lammel D.R., Barth G., Ovaskainen O., Cruz L.M., Zanatta J.A., Ryo M., de Souza E.M., Pedrosa F.O. Direct and indirect effects of a pH gradient bring insights into the mechanisms driving prokaryotic community structures. Microbiome. 2018;6:1–13. doi: 10.1186/s40168-018-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J., Kern D., German L., Mccann J., Martins G.C., Moreira A. Soil fertility and production potential. In: Lehmann J., Kern D.C., Glaser B., Woods W.I., editors. Amazonian Dark Earths: Origin, Properties, Management. Springer; Dordrecht: 2003. pp. 105–124. [Google Scholar]
- Leite D.C.A., Balieiro F.C., Pires C.A., Madari B.E., Rosado A.S., Coutinho H.L.C., Peixoto R.S. Comparison of DNA extraction protocols for microbial communities from soil treated with biochar. Braz. J. Microbiol. 2014;45:175–183. doi: 10.1590/s1517-83822014000100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth R.V. Least-squares means: the R package lsmeans. J. Stat. Software. 2016;69:1–33. [Google Scholar]
- Lindahl V., Bakken L.R. Evaluation of methods for extraction of bacteria from soil. FEMS Microbiol. Ecol. 1995;16:135–142. [Google Scholar]
- Lucena-Aguilar G., Sánchez-López A.M., Barberán-Aceituno C., Carrillo-Ávila J.A., López-Guerrero J.A., Aguilar-Quesada R. DNA source selection for downstream applications based on DNA quality indicators analysis. Biopreserv. Biobanking. 2016;14:264–270. doi: 10.1089/bio.2015.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C.C. The real dirt on rainforest fertility. Science. 2002;297:920–923. doi: 10.1126/science.297.5583.920. [DOI] [PubMed] [Google Scholar]
- Mathieson W., Thomas G.A. Simultaneously extracting DNA, RNA, and protein using kits: is sample quantity or quality prejudiced? Anal. Biochem. 2013;433:10–18. doi: 10.1016/j.ab.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Mendes L.W., de Lima Brossi M.J., Kuramae E.E., Tsai S.M. Land-use system shapes soil bacterial communities in Southeastern Amazon region. Appl. Soil Ecol. 2015;95:151–160. [Google Scholar]
- Merloti L.F., Mendes L.W., Pedrinho A., de Souza L.F., Ferrari B.M., Tsai S.M. Forest-to-agriculture conversion in Amazon drives soil microbial communities and N-cycle. Soil Biol. Biochem. 2019;137:107567. [Google Scholar]
- Meyer K.M., Klein A.M., Rodrigues J.L.M., Nüsslein K., Tringe S.G., Mirza B.S., Tiedje J.M., Bohannan B.J.M. Conversion of Amazon rainforest to agriculture alters community traits of methane-cycling organisms. Mol. Ecol. 2017;26:1547–1556. doi: 10.1111/mec.14011. [DOI] [PubMed] [Google Scholar]
- Mirza B.S., Potisap C., Nüsslein K., Bohannan B.J.M., Rodrigues J.L.M. Response of free-living nitrogen-fixing microorganisms to land use change in the Amazon rainforest. Appl. Environ. Microbiol. 2014;80:281–288. doi: 10.1128/AEM.02362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MO BIO Laboratories . 2016. MO BIO's PowerLyzer PowerSoil DNA Kit Handbook.https://www.qiagen.com/us/resources/resourcedetail?id=c98f64db-1db0-4163-a8d1-0f176c40f1c6&lang=en [Google Scholar]
- Moreira D. Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res. 1998;26:3309–3310. doi: 10.1093/nar/26.13.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A., Jain K., Shah A.R., Madamwar D. An efficient and cost-effective method for DNA extraction from athalassohaline soil using a newly formulated cell extraction buffer. Biotech. 2016;6:1–7. doi: 10.1007/s13205-016-0383-0. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento C., Homma A. EMBRAPA-CPATU; Belém: 1984. Amazônia: Meio ambiente e tecnologia agrícola. [Google Scholar]
- Navarrete A.A., Venturini A.M., Meyer K.M., Klein A.M., Tiedje J.M., Brendan B.J., Nüsslein K., Tsai S.M., Rodrigues J.L.M. Differential response of Acidobacteria subgroups to forest-to-pasture conversion and their biogeographic patterns in the western Brazilian Amazon. Front. Microbiol. 2015;6:1–10. doi: 10.3389/fmicb.2015.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete A.A., Diniz T.R., Braga L.P.P., Silva G.G.Z., Franchini J.C., Rossetto R., Edwards R.A., Tsai S.M. Multi-analytical approach reveals potential microbial indicators in soil for sugarcane model systems. PloS One. 2015;10 doi: 10.1371/journal.pone.0129765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves E.G., Petersen J.B., Bartone R.N., Da Silva C.A. Historical and socio-cultural origins of amazonian Dark Earths. In: Lehmann J., Kern D.C., Glaser B., Woods W.I., editors. Amazonian Dark Earths: Origin, Properties, Management. Springer; Dordrecht: 2003. pp. 29–50. [Google Scholar]
- Nielsen K.M., Calamai L., Pietramellara G. Stabilization of extracellular DNA and proteins by transient binding to various soil components. In: Nannipieri P., Smalla K., editors. Vol. 8. Springer; Berlin, Heidelberg: 2006. pp. 141–157. (Nucleic Acids and Proteins in Soil. Soil Biology). [Google Scholar]
- Novotny E.H., de Azevedo E.R., Bonagamba T.J., Cunha T.J.F., Madari B.E., Benites V.M., Hayes M.H.B. Studies of the compositions of humic acids from Amazonian Dark Earth soils. Environ. Sci. Technol. 2007;41:400–405. doi: 10.1021/es060941x. [DOI] [PubMed] [Google Scholar]
- Ogram A.V., Mathot M.L., Harsh J.B., Boyle J., Pettigrew C.A. Effects of DNA polymer length on its adsorption to soils. Appl. Environ. Microbiol. 1994;60:393–396. doi: 10.1128/aem.60.2.393-396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. 2018. Vegan: Community Ecology Package. R Package Version 2.5-1.https://CRAN.R-project.org/package=vegan [Google Scholar]
- Øvreås L., Forney L., Daae F.L., Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill M., McPartlin J., Arthure K., Riedel S., McMillan N.D. Comparison of the TLDA with the Nanodrop and the reference Qubit system. J. Phys. Conf. Ser. 2011;307 [Google Scholar]
- Paula F.S., Rodrigues J.L.M., Zhou J., Wu L., Mueller R.C., Mirza B.S., Bohannan B.J.M., Nüsslein K., Deng Y., Tiedje J.M., Pellizari V.H. Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol. Ecol. 2014;23:2988–2999. doi: 10.1111/mec.12786. [DOI] [PubMed] [Google Scholar]
- Pedrinho A., Mendes L.W., Merloti L.F., da Fonseca M.C., Cannavan F.S., Tsai S.M. Forest-to-pasture conversion and recovery based on assessment of microbial communities in Eastern Amazon rainforest. FEMS Microbiol. Ecol. 2019;95 doi: 10.1093/femsec/fiy236. [DOI] [PubMed] [Google Scholar]
- Portilho I.I.R., Savin M.C., Borges C.D., Tsai S.M., Mercante F.M., Roscoe R., de Carvalho L.A. Maintenance of N cycling gene communities with crop-livestock integration management in tropical agriculture systems. Agric. Ecosyst. Environ. 2018;267:52–62. [Google Scholar]
- Robe P., Nalin R., Capellano C., Vogel T.M., Simonet P. Extraction of DNA from soil. Eur. J. Soil Biol. 2003;39:183–190. [Google Scholar]
- Rodrigues J.L.M., Pellizari V.H., Mueller R., Baek K., Jesus E.C., Paula F.S., Mirza B., Hamaoui G.S., Tsai S.M., Feigl B., Tiedje J.M., Bohannan B.J.M., Nusslein K. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:988–993. doi: 10.1073/pnas.1220608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose-Amsaleg C.L., Garnier-Sillam E., Harry M. Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil Ecol. 2001;18:47–60. [Google Scholar]
- RStudio Team . Integrated development for R. RStudio, Inc.; Boston, MA: 2016. RStudio.http://www.rstudio.com/ [Google Scholar]
- Sagova-Mareckova M., Cermak L., Novotna J., Plhackova K., Forstova J., Kopecky J. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl. Environ. Microbiol. 2008;74:2902–2907. doi: 10.1128/AEM.02161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors–occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- Schriewer A., Wehlmann A., Wuertz S. Improving qPCR efficiency in environmental samples by selective removal of humic acids with DAX-8. J. Microbiol. Methods. 2011;85:16–21. doi: 10.1016/j.mimet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Sidstedt M., Jansson L., Nilsson E., Noppa L., Forsman M., Rådström P., Hedman J. Humic substances cause fluorescence inhibition in real-time polymerase chain reaction. Anal. Biochem. 2015;487:30–37. doi: 10.1016/j.ab.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Silver W.L., Neff J., McGroddy M., Veldkamp E., Keller M., Cosme R. Effects of soil texture on belowground carbon and nutrient storage in a lowland Amazonian forest ecosystem. Ecosystems. 2000;3:193–209. [Google Scholar]
- Stach J.E.M., Bathe S., Clapp J.P., Burns R.G. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. Ecol. 2001;36:139–151. doi: 10.1111/j.1574-6941.2001.tb00834.x. [DOI] [PubMed] [Google Scholar]
- Stahl D.A., Amann R. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons; Hoboken: 1991. pp. 205–248. [Google Scholar]
- Steward C. From colonization to “environmental soy”: a case study of environmental and socio-economic valuation in the Amazon soy frontier. Agric. Hum. Val. 2007;24:107–122. [Google Scholar]
- Terrat S., Christen R., Dequiedt S., Lelièvre M., Nowak V., Regnier T., Bachar D., Plassart P., Wincker P., Jolivet C., Bispo A., Lemanceau P., Maron P.A., Mougel C., Ranjard L. Molecular biomass and MetaTaxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure. Microb. Biotechnol. 2012;5:135–141. doi: 10.1111/j.1751-7915.2011.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermo Fisher Scientific . 2010. Nucleic Acid - Thermo Scientific NanoDrop Spectrophotometers.https://tools.thermofisher.com/content/sfs/brochures/Thermo-Scientific-NanoDrop-Products-Nucleic-Acid-Technical-Guide-EN.pdf [Google Scholar]
- Tsai Y.L., Olson B.H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl. Environ. Microbiol. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.L., Olson B.H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Agriculture (USDA) Soil Texture Calculator. https://www.nrcs.usda.gov/wps/portal/nrcs/detail/soils/survey/?cid=nrcs142p2_054167
- Valadares-Pereira A.A., Oliveira E.C.A.M., Navarrete A.A., de Oliveira Junior W.P., Tsai S.M., Peluzio J.M., de Morais P.B. Fungal community structure as an indicator of soil agricultural management effects in the Cerrado. Rev. Bras. Cienc. Solo. 2017;41 [Google Scholar]
- Wickham H. second ed. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wilson I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates C., Gillings M.R., Davison A.D., Altavilla N., Veal D.A. Methods for microbial DNA extraction from soil for PCR amplification. Biol. Proced. Online. 1998;1:40–47. doi: 10.1251/bpo6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C.C., Burghoff R.L., Keim L.G., Minak-Bernero V., Lute J.R., Hinton S.M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl. Environ. Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipper H., Buta C., Lämmle K., Brunner H., Bernhagen J., Vitzthum F. Mechanisms underlying the impact of humic acids on DNA quantification by SYBR Green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res. 2003;31:e39. doi: 10.1093/nar/gng039. [DOI] [PMC free article] [PubMed] [Google Scholar]