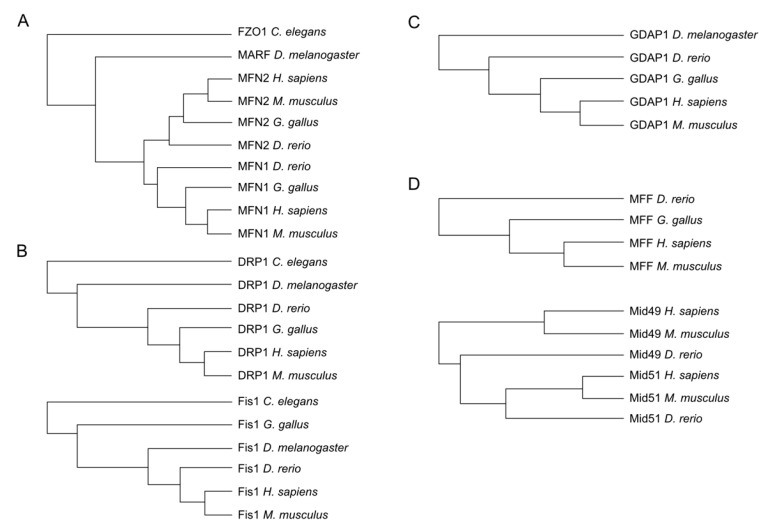
Figure 2.
Phylogenetic trees obtained by multiple alignment of amino acid sequences from Homo sapiens, Mus musculus, Gallus gallus, Danio rerio, and Drosophila melanogaster when possible. The most abundant isoforms were used for the alignment. (A) MFN2 is present in all compared species while MFN1 was not found in Drosophila and C elegans. (B) DRP1 and FIS1 presented orthologs in all compared species. (C) GDAP1 is present in all compared species except in C elegans. (D) MFF and MID49/51 proteins were found in Homo sapiens, Mus musculus, and Danio rerio.