Antiviral drugs have traditionally been developed by directly targeting essential viral components. However, this strategy often fails due to the rapid generation of drug-resistant viruses. Recent genome-wide approaches, such as those employing small interfering RNA (siRNA) or clustered regularly interspaced short palindromic repeats (CRISPR) or those using small molecule chemical inhibitors targeting the cellular “kinome,” have been used successfully to identify cellular factors that can support virus replication.
KEYWORDS: antiviral agents, host factors, drug resistance, epigenetic regulation, precision medicine
SUMMARY
Antiviral drugs have traditionally been developed by directly targeting essential viral components. However, this strategy often fails due to the rapid generation of drug-resistant viruses. Recent genome-wide approaches, such as those employing small interfering RNA (siRNA) or clustered regularly interspaced short palindromic repeats (CRISPR) or those using small molecule chemical inhibitors targeting the cellular “kinome,” have been used successfully to identify cellular factors that can support virus replication. Since some of these cellular factors are critical for virus replication, but are dispensable for the host, they can serve as novel targets for antiviral drug development. In addition, potentiation of immune responses, regulation of cytokine storms, and modulation of epigenetic changes upon virus infections are also feasible approaches to control infections. Because it is less likely that viruses will mutate to replace missing cellular functions, the chance of generating drug-resistant mutants with host-targeted inhibitor approaches is minimized. However, drug resistance against some host-directed agents can, in fact, occur under certain circumstances, such as long-term selection pressure of a host-directed antiviral agent that can allow the virus the opportunity to adapt to use an alternate host factor or to alter its affinity toward the target that confers resistance. This review describes novel approaches for antiviral drug development with a focus on host-directed therapies and the potential mechanisms that may account for the acquisition of antiviral drug resistance against host-directed agents.
INTRODUCTION
The recent report of the International Committee on Taxonomy of Viruses (ICTV; 2019) listed 4,958 viral species across 14 orders, 143 families, and 846 genera (1). Recent advances in technology are uncovering new viruses and/or their genetic/antigenic variants almost on a daily basis. The world is currently experiencing an outbreak of a new coronavirus disease (COVID-19) (2–5), but fortunately most new viruses are not associated with clinical disease. Throughout human history, some new viruses have been the cause of major epidemics (6–15), and today’s highly interconnected world makes us even more vulnerable than in the past. Whereas the most successful approach to control any virus infection is with vaccines, this strategy is not effective for many agents for a variety of reasons. A potentially more effective general approach to combat virus infections is to develop effective antivirals (16). This approach was first used to control a herpesvirus infection and is currently the major means of controlling human immunodeficiency virus type 1 (HIV-1) infection (17). Recently, highly effective antivirals were also developed to control hepatitis C virus (HCV) (18–20), and the world would welcome antivirals active against COVID-19.
The majority of the antiviral drugs that have been approved by the Food and Drug Administration (FDA) act by directly targeting virus-encoded factors (21). However, these drugs almost invariably lose efficacy due to the emergence of drug-resistant virus variants (22–27). Consequently, alternative antiviral approaches need to be explored. In this review, we make the case for developing antiviral drugs that target host factors needed by the virus but not mandatory for host cell functions. We refer to such drugs as host-directed antiviral agents. Two major approaches, genome-wide small interfering RNA (siRNA) and/or clustered regularly interspaced short palindromic repeats (CRISPR) screens and small molecule chemical inhibitors targeting the cellular “kinome,” have revealed those of the cell’s ∼25,000 protein-encoding genes that interact and regulate replication of the infecting virus (28, 29). Some of these host factors are dispensable for the host but are required by the virus to complete various steps of its life cycle (30–35). These host factors can serve as targets for antiviral drug development (Fig. 1). Since genetic variability of the host is quite low compared to viruses, host-directed antiviral agents are less likely to become ineffective because of mutations in the viral genome (36, 37), although examples of resistance to such agents have been described (38, 39). The major goals of this review are to discuss novel approaches of antiviral drug development, with a focus on host-directed antiviral agents. We also discuss potential mechanisms of drug resistance against host-directed antiviral agents.
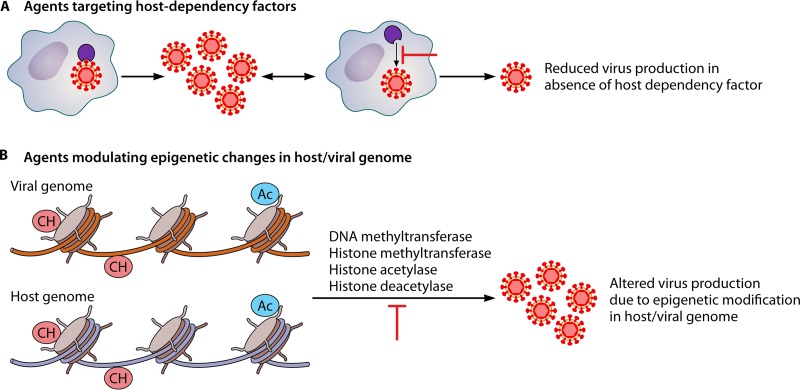
FIG 1.
Novel strategies of antiviral drug development. (A) In order to effectively replicate inside cells, virus is highly dependent on certain cellular factors, some of which are dispensable for cells and therefore may serve as targets for antiviral drug development. (B) Epigenetic changes such as DNA methylation and histone acetylation have also been shown to regulate viral replication/transcription/translation; thereby, inhibitors targeting the enzymes responsible for these epigenetic modifications (DNA methyltransferase, histone methyltransferase, histone acetylase, histone deacetylase) may serve as viable targets for antiviral drug development.
HOST-DIRECTED ANTIVIRAL AGENTS
Viruses establish numerous interactions with host factors and pathways during replication. In fact, technological advances have already identified several host factors that are essential for virus replication but dispensable for the host (40–42). Additionally, some host activities that respond to a viral infection, such as the interferon (IFN) and adaptive immune responses, can be manipulated to change the outcome of infection. Host-directed therapies have gained momentum in the past 2 decades. Many such studies are in the preclinical stages of development, with only a few compounds approved by the FDA. Herein we separately discuss FDA-approved drugs and therapeutic agents which are under preclinical development.
FDA-Approved Host-Directed Antiviral Agents
Of the majority of the antiviral drugs so far approved by the FDA, only a few are based on targeting host factors (Table 1) (21). Most of the FDA-approved host-directed antiviral drugs are based on IFNs and have been developed against chronic infections such as HIV-1, human papillomavirus (HPV), hepatitis B virus (HBV), and hepatitis C virus (HCV) (21) (Table 1). There are several different subtypes of IFNs, but so far only alpha IFNs (IFNs-α) are being used clinically (43). The main target for IFN therapy initially was treatment of non-A non-B chronic hepatitis (now known to be hepatitis C) (44). IFNs were later approved to treat HBV (45) and HPV (46) infections as well. Subsequently, an increased sustained virological response rate of IFN therapy was achieved by various technological modifications and the inclusion of ribavirin. The modifications included conjugation with polyethylene glycol to form pegylated IFN (PegIFN), which prevented enzymatic degradation, increased the half-life of IFNs, and also decreased, but did not eliminate, side effects (47–49). However, this approach still has many problems, such as patient unresponsiveness (50) and production of interfering (anti-IFN) antibodies (51), and has been largely discarded in the United States since the development of other, safer drugs.
TABLE 1.
FDA-approved host-directed antiviral agentsa
| Trade name | Generic name | Target virus | Mechanism of action | Yr |
|---|---|---|---|---|
| Intron A | IFN-α-2b | HPV | IFN mediated | 1988 |
| Alferon N injection | IFN-α-N3 | HPV | IFN mediated | 1989 |
| Condylox | Podofilox | HPV | Interrupts cell division cycle | 1990 |
| Intron A | IFN-α-2b | HCV | IFN mediated | 1991 |
| Intron A | IFN-α-2b | HBV | IFN mediated | 1992 |
| Infergen | Interferon alfacon-1 | HCV | IFN mediated | 1997 |
| Aldara | Imiquimod | HPV | Induction of cytokines | 1997 |
| Rebetron | PegIFN-α-2b plus ribavirin | HCV | IFN mediated | 1998 |
| Rebetol | Ribavirin | HCV | Multiple modes of action | 1998 |
| Pegintron/Sylatron | PegIFN-α-2b | HCV | IFN mediated | 2001 |
| Pegasys | PegIFN-α-2a | HCV | IFN mediated | 2002 |
| Pegasys | PegIFN-α-2a | HBV | IFN mediated | 2005 |
| Veregen | Sinecatechins | HPV | Immunomodulator | 2006 |
| Selzentry | Maraviroc | HIV-1 | Blocks gp120 and CCR5 interaction | 2007 |
IFN, interferon; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV-1, human immunodeficiency virus type 1; HPV, human papillomavirus; PegIFN-α, pegylated IFN-α.
Currently, PegIFN-α-2b and ribavirin combination therapy is recommended for the treatment of chronic HCV infection, which results in sustained viral clearance in more than 50% of patients (52). However, the precise mechanism of action of IFN‐α and ribavirin against HCV is not well defined. In fact, viral infection itself may suppress IFN‐α induction and action in nonresponders, as has been observed in animal models of HCV infection (53). Moreover, individual host genetic factors, innate and adaptive immune responses, and viral genetic diversity as well as coinfections may also account for part of the nonresponse to therapy (54). Alfacon-1 (recombinant synthetic type I interferon) and ribavirin proved effective in some patients who did not respond to standard PegIFN-α/ribavirin therapy (44). Currently, new drug combinations are under development. For instance, telaprevir (an inhibitor of the HCV NS3/4 protease) in combination with PegIFN-α/ribavirin can be effective in treating chronic HCV patients who are unresponsive to conventional PegIFN-α/ribavirin therapy (55). Several other HCV NS3/4A protease inhibitors used in combination with PegIFN-α and ribavirin may also achieve improved rates of a sustained virological response (56–59). However, the toxicity when they are combined with PegIFN-α and ribavirin still limits their overall efficacy (60, 61).
IFN preparations have several additional issues. These include fatigue, flu-like symptoms, neurological disorders, autoimmunity, ischemia, pneumonitis, anemia, neutropenia, nephritis, erythema, vasculitis, and necrosis, all of which limit their use (62–67). In addition, viruses resistant to IFN therapy have also been observed (68, 69); these may have emerged due to the acquisition of mutations in either structural or nonstructural proteins (70, 71).
In order to overcome the side effects of IFN therapy, targeting IFNs to local sites or preparing a prodrug formulation can be effective (72, 73). In this context, IFN-α-2 has been employed to target the liver with antibody specific to liver tissues (74, 75). Furthermore, to improve the pharmacokinetics and overcome pleiotropic effects, IFN-α-2b was engineered to make it latent by providing a protective shell of latency-associated protein of transforming growth factor β (TGF-β) fused with HCV NS3 protease cleavage site as a linker (67).
Host-directed antiviral drugs have also been approved to treat HPV-associated warts. In this context, Intron A (IFN-α-2b), a drug also approved to treat HCV and HBV infection, has shown clinical efficacy against HPV-associated genital warts (76). Another drug, podofilox (Condylox) potently inhibits cell mitosis, which eventually results in the regression of HPV-associated warts (77). In addition, imiquimod (Aldara) exhibits profound antitumor (antiwart) activity by acting on several immunological activities (78). Likewise, sinecatechins (Veregen) was the first botanical drug product approved by the FDA in 2006 for the treatment of external genital and perianal warts, although its mechanism of action is as yet not known. Another injectable preparation of IFN-α-N3 upregulates major histocompatibility complex class I (MHC-I) expression, which allows for enhanced presentation of virus-associated antigens, thereby activating cytotoxic CD8+ T cells that aid in wart regression (79).
Maraviroc, the only CCR5 antagonist licensed for clinical use, is a negative allosteric modulator of the C-C chemokine receptor type 5 (CCR5). Maraviroc inhibits HIV-1 entry by altering the extracellular loops of CCR5 in such a way that the HIV-1 gp120 envelope glycoproteins can no longer bind CCR5 (80, 81). In addition, maraviroc also reverses HIV-1 latency in vitro (82). However, HIV-1 strains with partial maraviroc resistance have appeared (83–85). The resistant strains may use alternate coreceptors (CXCR4 rather than CCR5) or alter their ability to bind the maraviroc-bound form of CCR5 (86–88).
Targeting Host Factors and Pathways Important for Viral Replication
Virus infection may activate intracellular signaling pathways, which regulate cell growth (37, 89–92), cell survival (37, 93), and/or immune activation (93, 94). In a typical course of virus infection, infected cells secrete IFNs, which induce nearby cells to express many so-called IFN-stimulated genes (ISGs), whose functions include antiviral activity. Viruses may also usurp intracellular signaling pathways to their own advantage in the cells they infect (37, 93). For example, in influenza A virus (IAV) infection, the nuclear factor kappa B (NF-κB) signaling pathway may be activated (95). This results in optimal synthesis of viral genomes, and it can also result in the secretion of proinflammatory cytokines (95, 96). Thus, signaling pathways such as NF-κB may be targeted by therapies to regulate both virus replication and the virus-induced inflammatory responses.
Blockade of cellular receptors or other cellular proteins that regulate virus replication has also been targeted by therapies, but so far very few have entered into clinical trials (21). In the majority of studies, kinases and lipid synthases are the major host targets for antiviral drug development (Table 2).
TABLE 2.
Host-directed antiviral agents under preclinical developmenta
| Host factor function | Antiviral agent(s) | Virus(es) | Host target | Reference(s) |
|---|---|---|---|---|
| Support viral entry | DAS181 | IAV, PIV | Sialic acid receptor | 405–411 |
| R448, cabozantinib | ZIKV | AXL kinase | 412, 413 | |
| Ezetimibe | HCV | NPC1L1 | 166 | |
| Obatoclax, chloroquine, bafilomycin A1, ammonium chloride, Arbidol (umifenovir), chlorpromazine, niclosamide, mefloquine HCl | ZIKV, FLV hRhV, IAV | Regulation of endosomal pH | 413–417 | |
| Glycyrrhizin | IAV | Regulation of endocytotic uptake | 418 | |
| Concanamycin, saliphenylhalamide | IAV | Cellular vacuolar ATPases | 419, 420 | |
| Daptomycin | ZIKV | Modulation of late endosomal function | 417 | |
| LJ001 | IAV, poxvirus, FLV, HIV-1 | Modulation of membrane fluidity | 421 | |
| Thapsigargin | PPRV, NDV | SERCA | 39 | |
| Dynasore | HSV-1 | Dynamin | 422 | |
| MLS000394177, MLS000733230, MLS000730532 | EBOV | Macropinocytic uptake | 423 | |
| Bisindolylmaleimide I, calphostin C, chelerythrine, enzastaurin, staurosporine | WNV, IAV | PKC | 424, 425 | |
| Fattiviracin | HIV-1 | Internalization factors | 426 | |
| Aprotinin, camostat | IAV | Protease inhibitor | 427, 428 | |
| Jasplakinolide, cytochalasin D | hAdV | Actin polymerization | 429 | |
| Amiloride (EIPA) | CVB3, hAdV | Sodium-proton exchange | 430 | |
| Emetine, cephaeline | PPRV, NDV, BPXV, BHV-1, ZIKV, EBOV | Lysosomal function | 431, 432 | |
| Tenovin-1 | ZIKV | SirT1 and SirT2 | 433 | |
| Clonidine | IAV | α2-Adrenergic receptors | 434 | |
| Nanchangmycin | ZIKV | AXL kinase | 433 | |
| Erlotinib | HCV | EGFR and GAK | 104, 165 | |
| Sunitinib | HCV | AAK1 | 104 | |
| NIM-811, Debio-025 | EV | Cyclophilin | 435 | |
| STI-571, Gleevec, imatinib, nilotinib, dasatinib | Poxvirus, PyV, HIV-1 | Abl family protein kinases | 436, 437 | |
| Support viral genome replication, transcription, and translation | SD-29 | HSV-1 | RACK1 | 438 |
| Torin1, rapamycin | CMV, BEFV | mTOR kinase | 439–441 | |
| Hippuristanol, silvestrol | CMV, ZIKV | eIF4A | 440, 442 | |
| CGP57380 | HSV-1, poxvirus, hCMV | MNK1 | 38, 443–445 | |
| 4E2RCat, 4EGI-1 | CoV, BPXV | eIF4E/eIF4G interaction | 38, 446 | |
| Apigenin | FMDV, EV71 | hnRNP A1 and A2 | 447, 448 | |
| AG879 | IAV, SV, HSV-1, MHV, RV | NGFR | 121, 129 | |
| Genistein | HIV-1 | Tyrosine kinase | 449 | |
| Tyrphostin A9 | IAV, SV, HSV-1, MHV, RV | PDGFR | 121, 129 | |
| Gefitinib (Iressa) | Poxvirus, hCMV | EGFR | 450, 451 | |
| Ivermectin | IAV | Importins | 452 | |
| Verdinexor, DP2392-E10, leptomycin B | IAV | XPO1 | 453–455 | |
| TG100572 | HSV-1 | Src family kinases | 156 | |
| Vemurafenib | IAV | Raf | 456 | |
| U0126, Cl-1040 (PD184352) | IAV, IBV, PEDV, AstV, BDV, CoV, JUNV, HSV-1 | MEK1/2 | 457–464 | |
| FR180204, Ag-126 | VEEV, DENV, lentivirus | ERK1/2 | 465, 466 | |
| SB203580 | EMCV | p38 | 467 | |
| AS601245, SP600125 | IAV, hCMV | JNK | 468, 469 | |
| Mycophenolic acid, ribavirin | ZIKV, IAV, RSV, CoV, EV71, CVB3, HCV | IMPDH | 417, 470–472 | |
| Leflunomide, compound A3 | FLV | DHODH | 368 | |
| TVB-2640 | HCV | FAS | 389, 473 | |
| Statins (atorvastatin, fluvastatin, lovastatin, pravastatin, and simvastatin) | HCV | HMG-CoA reductase | 393 | |
| Clypearin, corilagin, TG003 | IAV | CLK1 | 474 | |
| Silvestrol, pateamine | IAV | eIF4A | 475, 476 | |
| Curcumin, demethoxycurcumin, bisdemethoxycurcumin, EF-24, FLLL32 | JEV, RVFV, HCV, EV71 | Ubiquitin-proteasome, PKC, NF-κB, Akt | 477–480 | |
| PIK93, BF738735, GW5074, T-00127-HEV1 | EV | PI4KB | 481, 482 | |
| Itraconazole, 25-hydroxycholesterol, AN-12-H5, T-00127-HEV2, TTP-8307 | EV | OSBP | 481 | |
| EYP001 | HBV | Synthetic farnesoid X receptor | 483 | |
| APG-1387 | HBV | cIAP2 | 484 | |
| Fenretinide (4-HPR) | ZIKV, DENV | Activator of retinoid receptors | 485 | |
| Difluoromethylornithine, diethylnorspermine | ZIKV, DENV | Host polyamine synthesis | 486 | |
| Cyclopiazonic acid (CPA) | hRSV | Intracellular calcium ATPase | 487 | |
| MK2206 | IAV | Akt | 488 | |
| PD-0332991 | HSV-1 | CDK4/6 | 489 | |
| LDC4297 | hCMV, AdV | CDK7 | 490 | |
| JMN3-003 | Myxovirus | G1-phase arrest | 369 | |
| AGK2 | HBV | SirT2 | 491 | |
| Amiloride (EIPA) | CVB3, hAdV35 | Sodium-proton exchange | 429, 430 | |
| HL05100P2, cyclosporine, NIM-811, CRV431, CMX157 | EAV, PRRSV, HCV, HBV | Cyclophilin | 435, 492, 493 | |
| Emetine | PPRV, NDV, BPXV, BHV-1 | Unknown | 431, 432 | |
| Cephaeline | ZIKV, EBOV | Unknown | 431, 432 | |
| Nitazoxanide, tizoxanide | ZIKV, RV, NV, HBV, HCV, IAV | Unknown | 494 | |
| Glycyrrhizin | CVB3, hAdV, IAV | Unknown | 418, 429, 430 | |
| RG7834 | HBV | Unknown | 495 | |
| Veregen (sinecatechins) | HPV | Unknown | 21 | |
| Support virus assembly and release | Brefeldin A | DENV, HCV | ADP-ribosylation factor | 496, 497 |
| PF4620110, LCQ908 | HCV | DGAT1 | 498 | |
| AG879 | IAV, SINV, HSV-1, MHV, RV | NGFR | 121, 129 | |
| U18666A | IAV | Annexin A6 | 499 | |
| UV-4B | DENV, IAV | ER glycosylation pathway | 500 | |
| Tyrphostin A9 (A9) | IAV, SINV, HSV-1, MHV, RV | PDGFR | 121, 129 | |
| Verapamil, chlorpromazine | IAV, SINV, VSV | Calcium channel blocker | 501, 502 | |
| Gemfibrozil, lovastatin | IAV | Unknown | 503 | |
| Suramin | ZIKV | Glycosylation (secretory pathway) | 504, 505 | |
| Dynasore | HSV | Protein trafficking | 422 | |
| DEBIO-025 | HCV | Cyclophilin A | 506, 507 | |
| Bortezomib | ZIKV | Proteasome function | 417 | |
Abbreviations: AAK1, adaptor-associated protein kinase 1; AdV, adenovirus; AstV, astrovirus; BDV, Borna disease virus; BPXV, buffalopox virus; BHV1, bovine herpesvirus 1; BEFV, bovine ephemeral fever virus; CDK, cyclin-dependent kinase; CVB3, coxsackievirus B3; cIAP2, cellular inhibitor of apoptosis protein 2; CLK1, Cdc2-like kinase 1; CMV, cytomegalovirus; CoV, coronavirus; DGAT1, diacylglycerol acyltransferase-1; DENV, dengue virus; DHODH, dihydroorotate dehydrogenase; EAV, equine arteritis virus; EBOV, Ebola virus; EGFR, epidermal growth factor receptor; EMCV, encephalomyocarditis virus; EV, enterovirus; eIF4E, eukaryotic translation initiation factor 4E; ERK, extracellular-regulated kinase; FLV, flavivirus; FAS, fatty acid synthase; FMDV, foot-and-mouth disease virus; GAK, cyclin G-associated kinase; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV-1, human immunodeficiency virus type 1; HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; hnRNPA1, heterogeneous nuclear ribonucleoprotein A1; hRSV, human Rous sarcoma virus; hCMV, human cytomegalovirus; hAdV, human adenovirus; hRhV, human rhinovirus; HSV-1, herpes simplex virus 1; IAV, influenza A virus; IBV, influenza B virus; IMPDH, IMP dehydrogenase; JEV, Japanese encephalitis virus; JUNV, Junin virus; MHV, mouse hepatitis virus; MNK1, MAPK-interacting kinase 1; NDV, Newcastle disease virus; NGFR, nerve growth factor receptor; NPC1L1, Niemann-Pick C1-like 1; NF-κB, nuclear factor kappa B; OSBP, oxysterol-binding protein; PDGFR, platelet-derived growth factor receptor; PIV, parainfluenza virus; PEDV, porcine epidemic diarrhea virus; PI4KB, phosphatidylinositol 4-kinase IIIβ; PKC, protein kinase C; PPRV, peste des petits ruminants virus; PRRSV, porcine reproductive and respiratory syndrome virus; PyV, polyoma virus; RACK1, receptor for activated C kinase 1; RVFV, Rift Valley fever virus; RSV, Rous sarcoma virus; RV, rotavirus; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase; SV, Sendai virus; SINV, Sindbis virus; SirT1, Sirtuin type 1; VSV, vesicular stomatitis virus; VEEV, Venezuelan equine encephalitis virus; WNV, West Nile virus; XPO1, exportin 1; ZIKV, Zika virus.
Kinase inhibitors.
Completion of the Human Genome Project in 2002 identified 518 kinases that are collectively known as the cellular “kinome” (97). Kinases are implicated in various physiological processes to maintain cellular homeostasis, and their dysregulation could result in pathology. Infections by pathogens, including viral infections, are also associated with perturbation of the “kinome” (98, 99). Each step of the virus replication cycle can be regulated by multiple kinases (100–102). Some of the cellular kinases are dispensable for host cell viability but might be needed during virus infection (102). Such kinases represent potentially valuable drug targets (Fig. 1A). Kinase function can be inhibited by small molecule chemical inhibitors (103), and this approach is used in the cancer field. The topic has been extensively reviewed (104–120). Out of the hundreds of different kinase inhibitors developed so far, only 38 have been licensed for use as anticancer agents. Kinase inhibitor libraries have also been screened for antiviral activity, and some promising candidates have emerged as potential inhibitors of different steps of the viral life cycle (37, 38, 121–128).
The viral replication cycle is a multistep process that includes attachment, entry, genome synthesis, assembly of newly synthesized virion particles, and budding. Each step of the viral life cycle involves host cell kinases (Table 2), with a single kinase regulating one or multiple steps of the viral life cycle. For example, the mitogen-activated protein kinase (MAPK)-interacting kinase 1 (MNK1) inhibitor CGP57380 inhibits only initiation of buffalopox virus (BPXV) protein synthesis (38). In contrast, against IAV, the receptor tyrosine kinase (RTK) inhibitors AG879 and A9 can block multiple steps, including viral RNA synthesis, export of viral ribonucleoproteins (vRNP), and budding (121). While the precise mechanism by which RTK regulates viral RNA synthesis is not known, regulation of vRNP export and budding is mediated via the CRM1 (chromosomal maintenance 1) nuclear export pathway and the farnesyl pyrophosphate synthase (lipid biosynthesis enzyme), respectively (121). It is likely that targeting signaling pathways that regulate multiple steps of the viral life cycle will represent more effective antiviral approaches (121).
All members of a particular virus family usually share the same kinase requirements. For example, SERCA (sarco/endoplasmic reticulum calcium-ATPase) inhibitor blocks replication of multiple Paramyxoviridae family members (39). Nevertheless, some kinases are essential for multiple virus families and hence can represent potential targets for developing broad-spectrum antiviral drugs (121, 129). One such example is the RTK inhibitor AG879, which is active against IAV, rhesus rotavirus, Sendai virus, coronavirus, herpes simplex virus 1 (HSV-1), and Pichinde virus (an arenavirus) (121, 129).
Lipid biosynthesis inhibitors.
Besides nucleotides and amino acids, many viruses need a continuous supply of cellular fatty acids during their replicative cycle (130, 131). To achieve this, viruses may need to reprogram cellular metabolism, including lipid synthesis, to facilitate their own optimal replication (131, 132). Treatment of cells with chemical inhibitors that suppress fatty acid biosynthesis results in decreased virus production (121, 133–136), an approach that has shown promise against dengue virus (DENV), Zika virus (ZIKV), and West Nile virus (WNV) (137).
Acetyl coenzyme A (acetyl-CoA) carboxylase (ACC), ATP citrate lyase (ACLY), and fatty acid synthase (FASN) are known to regulate fatty acid biosynthesis in eukaryotic cells (138). Targeting ACC with the chemical inhibitor 5-(tetradecyloxy)-2-furoic acid (TOFA) and 3,3,14,14-tetramethylhexadecanedioic acid (MEDICA 16) has been shown to reduce flavivirus (WNV and Usutu virus [USUV]) replication (134). These compounds act by reducing cellular levels of multiple lipids, such as glycerophospholipids, sphingolipids, and cholesterol (134). Additionally, the lipid biosynthesis inhibitor TOFA and cerulenin exhibit broad-spectrum antiviral activity against ZIKV (Flaviviridae) and Semliki Forest virus (Togaviridae) by blocking ACC and FASN, respectively (133). Improved technologies, such as lipidomics, should provide insights into reprogramming of lipid metabolism following viral infections. However, caution is warranted, since targeting host lipid metabolism as an antiviral strategy may be limited by toxicity to host cells (139).
Besides targeting cellular kinases and fatty acid synthases, small molecule chemical inhibitors have also been developed against other protein/lipid targets, and these are summarized in Table 2. Other types of inhibitors include the relatively new class of therapeutic monoclonal antibodies (MAbs) that can be used to target certain host factors required for virus infection and replication. These are described in the following section.
Host-directed therapeutic monoclonal antibodies.
Rather than examining the role of antibodies directed against viral proteins, our focus in this section is to highlight the role of antibodies directed against host components when the net outcome is antiviral. For example, antibodies such as UB-421 (140), ibalizumab-uiyk (141), and maraviroc (80, 81), which block receptor binding sites on CD4+ T cells, have shown clinical efficacy in treating HIV-1 infection (21).
Cellular tight-junction proteins, such as claudin (142) and occludin (143), may act as entry receptors for viruses such as HCV (142–145). Thus, anti-claudin1 (CLDN1) (146) and anti-occludin (147) monoclonal antibodies have been designed and shown to inhibit HCV infection with minimal side effects. Similarly, the human scavenger receptor class B, type I (SR-BI), is a presumed receptor for HCV, and targeting this molecule may represent a useful therapeutic approach (148). However, HCV variants with decreased SR-BI dependency, which could limit the use of SR-BI targeting therapy, have been described previously (149–151). Interestingly, humanized mice infected with HCV variants that had increased in vitro resistance to SR-BI-targeting molecules still remain responsive to anti-SR-BI MAb therapy in vivo (152), hence representing an effective way to combat HCV infection (152).
Antibodies directed toward other cellular proteins (other than viral receptors) have also been developed. For example, blocking CD69 (a transmembrane C-type lectin protein in the host that is highly expressed by leukocytes upon infection) using anti-CD69 monoclonal antibodies can increase leukocytic numbers in secondary lymphoid organs during infection. This promotes clearing of vaccinia virus infection (153) (Fig. 2A). Anti-CD69 also increases the numbers of gamma interferon (IFN-γ)- and tumor necrosis factor alpha (TNF-α)-producing natural killer (NK) and adaptive immune T cells, an effect mediated in part by mTOR signaling (154, 155). Other studies have documented the therapeutic potential of bevacizumab (a recombinant humanized monoclonal antibody directed against vascular endothelial growth factor) to inhibit HSV-1-induced corneal neovascularization and also scarring in herpetic stromal keratitis (156, 157).
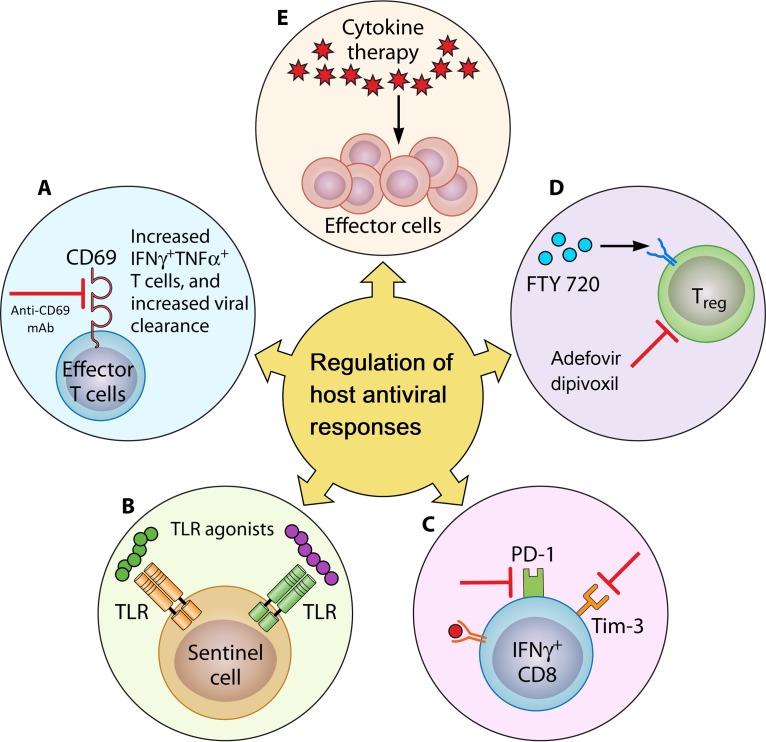
FIG 2.
Regulation of host antiviral responses. An antiviral immune response can be boosted by several possible means. (A) Monoclonal antibodies. Blocking CD69 by using anti-CD69 monoclonal antibodies increases leukocytic numbers in the secondary lymphoid organs during infection and improves the capacity to clear viral infection. (B) PRR agonists. Agonists targeting PRRs are another possible strategy to potentiate the innate immune response for enhanced virus clearance. (C) Modulation of counterinflammatory mechanisms. Counterinflammatory mechanisms such as the Tim-3/Galectin-9 interaction and the PD-1/PDL-1 axis prevent collateral tissue damage caused by an excessive immune response. Thus, antiviral immunity can be augmented by blocking these counterinflammatory mechanisms. (D) Manipulating Treg responses. Treg are the suppressor cells that act to limit an excessive immune response. FTY720 expands and potentiates Treg function, which in turn ameliorates virus-induced immunopathology. On the other hand, inhibiting Treg (adefovir dipivoxil) enhances antiviral effector responses. (E) Cytokine therapy. Administration of proinflammatory cytokines and alternatively a blockade of immunosuppressive cytokines may serve to enhance antiviral immune responses.
Monoclonal antibodies have been used as adjunct therapeutic agents along with antiviral therapy. For instance, oral anti-CD3 antibody therapy can cause significant reductions in viral load and an increase in regulatory T-cell levels in chronic HCV patients who additionally receive IFNs plus ribavirin therapy (158).
It is noteworthy that host-directed antiviral therapies (e.g., monoclonal antibodies and other types of inhibitors) offer several advantages over virus-directed interventions. Compared to virus-directed agents that exert genotype-dependent antiviral activity, host-directed agents show broad-spectrum pan-genotypic activity. For example, monoclonal antibodies directed against SR-BI (159, 160), CLDN1 (161), and CD81 (162, 163) have shown broad-spectrum antiviral efficacy against multiple HCV genotypes. Likewise, host-directed agents such as ITX-5061 (164), erlotinib (165), ezetimibe (166), flavonoids (167, 168), lectins (169), and phosphorothioate oligonucleotides (170), as well as silymarin (171, 172), exhibit antiviral activities against multiple genotypes of HCV. Host-directed agents also restrict replication of viral escape variants. For example, besides blocking entry of all major HCV genotypes, monoclonal antibodies directed against CLDN1 can also inhibit cell entry of highly infectious neutralizing antibody escape variants of HCV (161). As a result, recent developments of clinically useful monoclonal antibodies and other host-directed agents have revolutionized strategies for antiviral therapy.
Regulating Host Antiviral Responses
An alternative to using drugs that directly target either viral events or physiological processes involved in replication in infected cells is to target the various host immune events set into play by viral infections. In response to infection, viruses can trigger a wide range of host responses that usually act to eventually control the extent of the infection and remove virus from the host (Fig. 2). Such events can be changed by an expanding series of therapies to facilitate host-directed viral control and to limit the extent of tissue damage caused by the infection. These strategies are discussed in the following sections.
Induction of the IFN pathways.
Viruses themselves may possess one or more pathogen-associated molecular patterns (PAMPs), which can be proteins, lipids, or nucleic acids. These PAMPs interact with cellular pattern recognition receptors (PRRs) to induce innate immune events such as IFN production (173–178). Cells respond to IFNs by changing the expression of a multitude of cellular proteins, collectively known as IFN-stimulated genes (ISGs) (179, 180). These function to protect the responding cells from viral replication and therefore help to resolve infections (181, 182). Other molecules produced in response to viral infection may participate in the cellular inflammatory response, and these too can exert antiviral control, although if overstimulated may contribute to tissue damage, as is typical during many chronic viral infections (183, 184).
The best-studied examples of viral PAMPs are those which trigger the cellular Toll-like receptors (TLR). Such interactions result in a cascade of events that include cell activation, the production of cytokines, and several other activities that can modulate the outcome of viral infection (185, 186) (Table 3). The PRRs may be triggered by synthetic ligands. For instance, administration of the TLR7 agonist GS-9620 may expand NK cells and HBV-specific T cells, which leads to better control of chronic HBV infection (187) (Fig. 2B). Likewise, the TLR7/8 agonist R848 can block ZIKV genome and protein synthesis in human monocytes via the activation of viperin, an antiviral protein (188). Similarly, TLR9 and TLR3 agonists can inhibit HCV and HBV replication, respectively (189).
TABLE 3.
Regulation of host antiviral responsesa
| Antiviral agent(s) and functional category | Virus(es) | Host target | Reference(s) |
|---|---|---|---|
| Agonist of the innate immune receptors | |||
| Rintatolimod (Ampligen) | HIV, HCV, HBV | TLR3 agonist | 189 |
| GS9620, RO6864018, RO7020531, AL-034, imiquimod (Aldara) | HBV, HPV | TLR7 agonist | 21, 508 |
| GS9688 | HBV | TLR8 agonist | 508 |
| CL097 | HIV | TLR7/8 agonist | 189 |
| PF-04878691 or 852A | HCV | TLR7/8 agonist | 189 |
| CPG10101, IMO-2125, SD-101 | HCV | TLR9 agonist | 189 |
| Inarigivir (SB 9200) | HBV | RIG-I agonist | 508 |
| Regulation of inflammatory pathway | |||
| IFN-α, PegIFN-α, Alferon N | HPV, HCV, HBV | TNF-α-mediated antiviral activity | 21 |
| Quercetin | JEV, HCV | TNF-α-mediated antiviral activity | 509–511 |
| A23187 | SINV, VSV | Ca2+ efflux-mediated antiviral immunity | 512 |
| Phorbol myristate acetate | HBV | Synthesis of NF-κB-mediated antiviral protein | 513, 514 |
| CI1033 | Variola virus | ErbB1(EGFR)-mediated antiviral immunity | 515 |
| SIP agonist | IAV | Suppression of virus-induced cytokine storm | 516, 517 |
| COX-2 depletion | IAV | Induction of type I IFN-mediated antiviral immunity | 518 |
| Statins | IAV | Anti-inflammatory and immunomodulatory effects | 519, 520 |
| PPAR agonist | IAV | Suppression of virus-induced cytokine storm and associated lethality in mice | 521–524 |
Abbreviations: COX-2, cyclooxygenase 2; DHV, duck hepatitis virus; EGFR, epidermal growth factor receptor; HBV, hepatitis B virus; HCV, hepatitis C virus; HPV, human papillomavirus; IAV, influenza A virus; JEV, Japanese hepatitis virus; PPAR, peroxisome proliferator-activated receptor-γ; SINV, Sindbis virus; SIP, sphingosine-1-phosphate; VSV, vesicular stomatitis virus.
Unlike cellular RNA, some viral RNAs contain a 5′ triphosphate (5′ppp) terminal structure. This is sensed by the cellular retinoic acid-inducible gene I (RIG-I), a member of the cytosolic PRR family that also activates intracellular signaling cascades to induce proinflammatory cytokine responses (190, 191). Administration of synthetic 5′ppp RNA may also activate RIG-I-dependent antiviral responses to provide significant protection against a diverse group of RNA and DNA viruses (192).
Certain drugs and biologicals that stimulate IFNs have been developed, and these have contributed to controlling virus infection. For instance, virus replication is suppressed by hydroxyquinolines, a class of small molecule compounds that activates IFN regulatory factor 3 (IRF3) of the type I IFN pathway (193).
Augmentation of host antiviral responses by inhibiting counterinflammatory pathways.
Host counterinflammatory pathways function to inhibit the collateral tissue damage that might occur as a consequence of excessive immune responses generated against a viral infection. On the other hand, host counterinflammatory mechanisms also dampen effective immunity to acute viral infections. For example, in a mouse model of IAV infection, a TIM-3/Galectin-9 immunoinhibitory interaction can act to limit collateral damage by inducing apoptosis of TIM-3-positive CD8+ T cells that mediate the damage (194). However, unfortunately, the approach can impair antiviral control, which is also mediated by CD8+ T cells. Accordingly, blocking the TIM-3/Galectin-9 immunoinhibitory interaction using TIM-3 fusion protein can enhance antiviral immunity by generating a more robust acute-phase virus-specific CD8+ T-cell response as well as increased levels of virus-specific serum IgM, IgG, and IgA antibodies (194) (Fig. 2C).
Other immune-directed approaches include blockade of the coinhibitory receptor programmed death-1 (PD-1). This has been used to control chronic simian immunodeficiency virus (SIV) infection in macaques with antibody to PD-1. This enhances protection via effects on virus-specific CD8+ T-cell function (195) (Fig. 2C). Others have also reported the benefits of controlling virus infection using MAbs that target inhibitory receptors, and the approach seems to have a promising future (196).
Another potential immune-directed strategy to combat virus infections is to target the function of regulatory T cells (Treg), which often act to diminish excessive inflammatory responses. Although Treg have beneficial effects against inflammatory reactions to viruses, the downside is potentially limiting protection, especially to acute infections (197). Thus, Treg activity needs to be therapeutically managed to potentiate antiviral immunity (Fig. 2D). For example, an acyclic nucleotide analogue of adenosine used against chronic HBV infection (198) inhibits Treg function as well as its expansion (199) but is mildly nephrotoxic. Similarly, TNF-α inhibited the suppressive effect of Treg, and this resulted in enhanced HBV-specific immune responses (200). However, inhibition of virus‐specific Treg can be a technically challenging issue for various reasons, as described in a recent review article (201). Besides directly regulating antiviral responses, manipulation of Treg can also ameliorate the tissue damage caused by viral infections. For example, long-term application of the sphingosine-1 phosphate receptor agonist FTY720 results in anti-inflammatory effects (202) in HSV-1-induced immunopathology. These effects of FTY720 were mediated by the conversion of T-cell receptor (TCR)-stimulated nonregulatory CD4+ T cells to Treg with increased suppressive activity. A plethora of inhibitory pathways of the immune system are important for maintaining self-tolerance and minimizing collateral tissue damage. It is also well understood that viruses coopt certain immune checkpoint pathways as a major mechanism of immune resistance, particularly against T cells that are viral antigen specific. Immune checkpoint modulation is thus a viable emerging treatment modality. However, careful consideration should be exercised before any treatment decisions are made.
Cytokine therapy.
Attempts have been made to enhance the therapeutic effect of antiviral adaptive immunity by administering cytokines. There are three phases of the T-cell immune response: expansion, contraction, and memory (203, 204). Interleukin 2 (IL-2) therapy during the contraction phase of an antiviral immune response can result in enhanced proliferation and survival of virus-specific T cells as well as a drastic reduction in viral titers (205) (Fig. 2E). However, IL-2 treatment during the expansion phase had a negative impact on the survival of rapidly dividing effector T cells (205). Similarly, in chronic HBV patients, the virus can be controlled using therapies that stimulate certain cytokines. For example, Peg-IFN-λ treatment achieves control likely by inducing high serum levels of IL-18 (antiviral cytokine) along with sustaining both IFN-producing HBV-specific CD4+ T cells and CD8+ T-cell responses (206). These enhanced immune activities can severely restrict virus replication (13). On the other hand, immunosuppressive cytokines, such as IL-10, can suppress antiviral immunity, and overcoming this effect can result in enhanced viral control, as has been observed with lymphocytic choriomeningitis virus (LCMV) infection (207).
Regulation of cytokine storm.
The term “cytokine storm” is used to describe aberrant production of cytokines and the tissue-damaging immunopathology they orchestrate. Originally, “cytokine storm” was coined to characterize pathology associated with organ transplantation, as demonstrated by an inflammatory response triggered by the donor immune cells reacting to the recipient patient host’s tissues (graft-versus-host disease) (208). Cytokine storm has also been correlated with increased disease severity, particularly in cases of acute viral infections (208). These include IAV (209, 210), DENV (211, 212), and Ebola virus (EBOV) (213) infections. Over 150 cytokines may be involved in a cytokine storm (211–213), but those primarily involved include TNF-α, IL-6, and IFNs.
Controlling cytokine storms by targeting host proteins involved in the activation of cellular signaling pathways is a potential approach to dampen tissue damage (214). For example, the highly pathogenic H5N1 avian influenza virus induces a robust cytokine response in comparison to seasonal flu (H1N1) (215). In addition, the highly pathogenic “Spanish flu” (H1N1 IAV strain), which caused a catastrophic pandemic in 1918-1919, was also shown to induce hypercytokinemia in ferrets (216). Since hypercytokinemia involves multiple cytokines, disrupting a single cytokine usually has limited value (215, 217). However, since the induction of the majority of the cytokines is mediated via activation of the NF-κB signaling pathway, targeting this transcription factor may be a therapeutically viable approach. For example, in a knockout mouse model of H5N1 infection, depletion of NF-κB (p50 subunit) resulted in a drastic reduction in the expression of the NF-κB-regulated cytokines and chemokines (lack of hypercytokinemia) (215, 218).
In one highly pathogenic IAV infection (H5N1), the majority of patients who experienced a cytokine storm were elderly or had compromised immune systems (219). In fact, a relevant question is why certain individuals are relatively resistant to cytokine storms whereas others are more susceptible. Hyper- and hyporesponders to bacterial products have also been identified in patients, which can be explained in part by differences in structure and function of their TLR1 proteins (220). Thus, the association between high host susceptibility to virus-induced cytokine storms and genetic polymorphisms in the PRRs needs to be explored in more detail.
Modulating Epigenetic Modifications
Epigenetics is the study of phenotypic changes which do not involve nucleotide variations in the genome of the organism (221). Major epigenetic changes that take place in cells are due to histone modifications (acetylation and methylation), phosphorylation, ubiquitination, and sumoylation. Histones interact with genomic DNA to form chromatin structures. The level of chromatin compaction depends primarily on methylation and/or acetylation of the histone proteins (222), and this determines genomic stability, gene expression, cell lineage development, stem cell maturation, and mitosis (223). Histone modifications are carried out by epigenetic regulators, such as histone acetyltransferases (HATs), histone deacetylases (HDACs), and histone methyltransferases (HMTs), and these factors have been targeted to treat cancers (224) and parasitic diseases (225). Likewise, DNA methyltransferases (DNMTs), which regulate DNA methylation, have been associated with many different diseases (226, 227). Recent reports also highlight the involvement of these epigenetic modifiers in regulating virus replication (228–231). For instance, epigenetic modifications can serve as an antiviral defense mechanism by suppressing transcription and replication of the viral genome. On the other hand, viruses may also epigenetically modulate host functions by inducing DNA hypermethylation of the host genome upon virus infection (232). Therefore, ongoing efforts to develop inhibitors of epigenetic regulators (Fig. 1B) to control gene expression during viral infection are warranted.
Histone acetylation and deacetylation.
Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminal tail that protrude from the histone core of the nucleosome are acetylated and deacetylated during gene regulation (233). Histone acetylation is regulated by HATs and HDACs, and chemical inhibitors of HDACs have been evaluated for antiviral effects. For example, the selective HDAC6 inhibitor tubacin has been shown to block Japanese encephalitis virus (JEV) (234) and HCV (235) replication. The mechanism involved induction of heat shock protein 90 (Hsp90; an HDAC6 substrate) hyperacetylation that inhibits Hsp90 and JEV NS5 interaction to block viral RNA synthesis (234). Similarly, the HDAC inhibitor SAHA (suberoylanilide hydroxamic acid) can inhibit HCV replication by increasing H3 acetylation levels (236) of an immunomodulatory protein, osteopontin (237).
In an in vitro model of latent HSV-1 infection, treatment with trichostatin A (TSA) and sodium butyrate (HDAC inhibitors) of quiescently infected PC12 cells (rat neuroblastoma) was shown to reactivate virus replication (238), suggesting that epigenetic modifications may be exploited to understand the issue of viral latency. Like other herpesviruses, Epstein-Barr virus (EBV) persists mainly as an episome (also called a covalently closed circular DNA [cccDNA]) in latently infected B lymphocytes (239). During latency, viral DNA is maintained with relatively low levels of histone acetylation. Expression of the viral BZLF1 protein is associated with viral lytic reactivation (240). Its expression is blocked by the recruitment of cellular repressive factors such as YY1 (Yin Yang 1) and ZEB (zinc finger E-box-binding factor). These prevent access of transcriptional activators and facilitate binding of repressive cofactors such as HDAC (240). Therefore, HDAC inhibitors can induce reversal of EBV latency.
Although these HDAC inhibitors have the potential for use as promising therapeutic agents, a careful assessment prior to their use is essential, as they have the potential to reactivate other latently infected viruses in the host’s genome (241). For instance, administration of SAHA/TSA can increase the severity of coxsackievirus B3 (CVB3)-associated myocarditis (242).
Controlling chronic HBV infection, which represents a major problem, might be achieved by manipulating epigenetic events in chronically infected cells. HBV genomes in viral particles remain in a circular, partially double-stranded DNA conformation that is transcriptionally inert (243). To become transcriptionally active, it is converted into cccDNA (episome) in the nuclei of infected cells (244, 245). Once infection has occurred, the virus persists indefinitely in the nuclei of hepatocytes as cccDNA. Currently approved antiviral therapy, based on nucleoside analogs, targets cytoplasmic HBV genomic replication without directly affecting cccDNA and therefore necessitates long-term antiviral therapy (246–250). Total cure of chronic HBV can be achieved only if the viral episomes are removed from the nuclei of all hepatocytes. The formation of cccDNA by HBV is facilitated by the cellular transcriptional machinery (251), which also involves epigenetic modifications (251, 252). The epigenetic inhibitor C646 (HAT inhibitor) transcriptionally silences cccDNA without any measurable toxicity, thus representing a novel approach to the therapy of chronic HBV infection (253).
Histone methylation.
HMTs are of two types, histone-lysine N-methyltransferases and histone-arginine N-methyltransferases. HMT inhibitors have proven their therapeutic potential against certain types of cancer, and they might be effective in regulating some viral infections. For example, PRMT5 (protein arginine methyltransferase 5) restricts HBV replication, which is mediated via epigenetic repression (demethylation of arginine residues at the arginine-rich C-terminal domain) of viral DNA transcription (254). Thus, PRMT5 agonists are valuable for repressing HBV DNA transcription (255).
Another potential use of HMTs is to manipulate HSV latency. The transcriptional repressor CTCF, also known as CCCTC DNA binding factor or 11-zinc finger protein, is a transcriptional factor in the human genome encoded by the CTCF gene. This can bind to HSV-1 DNA and promote HSV-1 lytic transcription. CTCF depletion by siRNA can cause an increase in repressive histone marks H3K27me3 and H3K9me3, concomitant with decreased transcription of HSV-1 genes (256). Likewise, treatment with 5′-deoxy-5′-methylthioadenosine (MTA; a protein methylation inhibitor) can suppress the level of H3K4me3 (mediated via methyltransferase Set1), which eventually results in reduced HSV-1 transcription and replication (257).
Histone demethylation.
DNA viruses encapsidate their genomes without histones but rapidly acquire chromatin structure following infection (258, 259). Therefore, they are subjected to modification by the host epigenetic modifiers, which can be inhibited to regulate viral replication. For example, inhibition of lysine-specific demethylase 1 (LSD1; KDM1A family) results in inhibition of transcription and lytic replication of the DNA viral genome. This results in reduced virus shedding and decreased disease severity (260–264). Unlike DNA viruses, RNA virus genomes do not depend on histone association and chromatin structure. However, surprisingly, LSD1 indirectly inhibits IAV replication by demethylating and activating interferon-induced transmembrane protein 3 (IFITM3), which serves as a cellular antiviral protein for IAV and several other RNA viruses (265). Additionally, even though HIV-1 is an RNA virus, blocking LSD1 activity has also been shown to suppress the proviral DNA activation of HIV-1 transcription in latently infected T cells. This effect is mediated by demethylation of the viral accessory Tat protein (266).
DNA methylation.
DNA methylation is a well characterized epigenetic modification that is associated with many different diseases, including microbial infections and cancers (226, 227). DNA methylation is most dynamic in CpG islands near transcription start sites (222). CpG islands are typically hypomethylated in comparison to the rest of the genome. Generally, promoter methylation represses gene transcription, while methylation at other regions of the genome induces gene transactivation (222). The chromatin structure undergoes changes following DNA methylation (267, 268) and during interaction with HDAC/DNMTs (269, 270). DNMTs bind the methyl group (-CH3) at the carbon 5 position of cytosine in CpG dinucleotides to methylate DNA (271). Hypermethylated DNA, which negatively correlates with hypoacetylated histones, leads to chromatin condensation and hence transcriptional repression (267).
By suppressing transcription and replication of the viral genome, DNA methylation serves as an antiviral defense mechanism. Endogenous retroviruses and retrotransposons are well known to be repressed by DNA hypermethylation (272). The genome of DNA viruses such as HPV, HSV-1, HBV, EBV, and adenovirus also undergo abundant methylation and are often silenced in the infected cells (273–277). The silenced viral genomes can be reexpressed by manipulating epigenetic events. For example, treatment with 5-azacytidine (decitabine), a DNMT inhibitor, induced activation of silenced EBV genomes. This eventually facilitated immune-mediated destruction of EBV-associated tumor cells (278, 279). However, demethylating agents can also activate retroviral RNA transcription from its dormant (latent) stage (280), a potential problem for therapy.
Viruses may also epigenetically subvert host functions by inducing DNA hypermethylation (281, 282). For example, HCV infection induces DNMT1 and 3b-mediated DNA hypermethylation. This leads to downregulation of expression of E-cadherin, a primary cell adhesion molecule with tumor suppressor activity (283). The effect can be reversed following treatment with a DNMT-specific inhibitor (283). Similarly, methylation of SOCS1 (suppressor of cytokine signaling 1, a negative regulator of JAK/STAT signaling) negatively correlated with HBV-induced hepatocellular carcinoma (284). Likewise, the promoter of GADD45 (growth arrest and DNA damage-inducible gene 45), a tumor suppressor gene, has been shown to be hypermethylated during HCV infection in mice (285). By regulating multiple DNMTs, DNA tumor viruses such as HPV and HBV induce aberrant DNA methylation (286–288), which can eventually result in carcinogenesis (289).
Techniques such as photo-cross-linking-assisted m6A sequencing (PA-m6A-seq), high-resolution mapping of N6-methyladenosine using m6A cross-linking immunoprecipitation (m6A-CLIP), bisulfite sequencing, 5-methylcytosine RNA immunoprecipitation (m5C-RIP), 5-azacytidine-mediated RNA immunoprecipitation (Aza-IP), and methylation-individual nucleotide resolution cross-linking immmunoprecipitation (miCLIP) have all emerged as powerful tools to profile epigenetic modifications in viral and host genomes (290–294). These techniques, when combined with chromatin immunoprecipitation (ChIP), can provide comprehensive information on the multiprotein complexes associated with epigenetic control of viral infections. In addition, elucidating the role of other modes of posttranslational modifications, such as phosphorylation or sumoylation, can provide insights into dynamic virus-host interactions (294).
MODERN APPROACHES OF ANTIVIRAL THERAPY AND TO IDENTIFY HOST FACTORS FOR ANTIVIRAL DRUG DEVELOPMENT
Drug Combination Approach
The appropriate use of drug combinations may result in enhanced potency and broadened antiviral activity and may lessen the chance of drug resistance (295, 296). As discussed above, the addition of ribavirin to PegIFN-α-based regimens provided dramatic improvement of chronic HCV control (53, 297). However, the mechanism by which IFN-α and ribavirin act against HCV has not been elucidated. Other combinations have also proven effective against HCV. For example, Xiao and colleagues showed that a combination of host-directed agents (erlotinib, dasatinib), host-directed antibodies (anti-CLDN1, anti-CD81, and anti-SR-BI), and virus-directed agents (telaprevir, boceprevir, and simeprevir or danoprevir, daclatasvir, mericitabine, and sofosbuvir) were highly effective against HCV (298).
A new avenue in the use of drug combinations may be to simultaneously target multiple host factors and pathways that are supportive of virus replication (299, 300). It is an added advantage if the host-directed agents have already been FDA approved. Such drugs can be used immediately to treat viral infections, an approach known as drug repurposing (299, 301–303). Because the targets of these drugs have already been well characterized and validated, they present with little to no safety issues and risks. To determine which drugs might be appropriately combined to exploit their different mechanisms of action, a number of new approaches have been tried and are discussed below.
Antiviral Drug Development in the Era of Precision Medicine
There is a significant gap in our understanding about how different individuals develop disease and respond to treatments. Traditional medicine is based on the “one-size-fits-all” approach, and this may miss its mark, because each person’s genetic makeup is slightly different from that of others. The promising idea of precision medicine (also known as personalized medicine, individualized medicine, or genomic medicine) is to cater healthcare to each person’s unique genetic makeup. However, this new technology is still being validated and may take several years to become clinically feasible. Regardless, it is important to consider the individual’s uniqueness when developing antiviral drugs, as this uniqueness can have an important impact on the outcome of therapies.
Patient-to-patient variability in the outcome of antiviral therapies might be due to different genetic profiles as well as the variable microbiome present in the individual (304, 305). The latter could influence host pharmacokinetic and pharmacodynamic profiles as well as drug distribution (306). The advent of methodologies to comprehensively characterize patients at genomic, transcriptomic, proteomic, metabolomic, and lipidomic levels along with the availability of computational tools to analyze global protein-protein interactions (interactome) has greatly improved biological databases (307). Employing these “omics” approaches to classify clinical populations into mechanistic subgroups is likely to result in a higher success rate of treatment modalities, including antiviral therapies (306).
One potential application of precision medicine is to predict the outcome of viral infections in different individuals. For example, IAV infection may induce robust proinflammatory cytokine responses, with some individuals experiencing severe disease. Under such circumstances, steroids are usually not advisable because they may promote virus replication (308). Furthermore, anti-inflammatory therapy aimed to dampen inflammatory responses has been successful but only in a few patients (308). Under such circumstances, the availability of “omics” data (biomarkers) should prove valuable to explain individual variations, to monitor immune responses, and to assess disease severity in individual patients.
Another application might apply to HIV-1 control. In HIV-1, restoration of CD4+ T-cell numbers is critical after initiation of antiretroviral therapy (ART) (309). However, it is evident that not all HIV-1 patients experience a rebound in their CD4+ T-cell counts, and they are therefore vulnerable to opportunistic infections (309, 310). Determining the genetic factors which predict different levels of patients’ responses to ART is key to optimal treatment outcomes. For example, the pattern of gene expressions by peripheral blood monocytes (PBMCs) of an individual patient can predict if certain individuals recover their CD4+ T-cell counts (311). Similarly, a polymorphism in the MDR-1 gene in patients has been associated with more potent responses to ART than of patients lacking certain polymorphisms (312–315). Polymorphisms in genes encoding drug transporters and metabolic enzymes are also important factors in determining concentrations of antiretrovirals in plasma (316–320). It is noteworthy that besides host genetic factors, alterations in the microbiome of individual patients may also influence the outcome of viral infections or vaccination (304). This is an important topic that has been discussed elsewhere (304, 321).
Genome-Wide Screens To Identify Host Factors for Drug Development
Classical reductionist approaches by studying the role of a single or a limited number of proteins do not provide a holistic view of all the cellular factors that can support virus replication. Modern drug development has been fueled by the advent of high-throughput genome-wide technologies, such as RNAi and CRISPR screens, which permit the simultaneous evaluation of multiple molecular targets. Data generated from the new high-throughput approaches have proven to be invaluable in assembling novel hypotheses and in identifying new and useful diagnostic biomarkers.
siRNA screens.
Small interfering RNAs (siRNAs) are artificially synthesized, 19- to 23-nucleotide-long double-stranded RNA molecules designed to specifically target a cellular mRNA for degradation (322). Through genome-wide RNAi screening assays, thousands of distinct host factors have been shown to either facilitate or inhibit replication of a variety of viruses (323–350). It is, however, less well understood how the majority of these cellular factors may impact the life cycle of different viruses. For instance, a total of 1,362 virus-supportive host factors have been identified in seven different IAV RNAi screens performed to date. The overlapping genes were shown to regulate various steps of the virus replication cycle (351–355). However, only a few of the thousands of identified host factors, such as NF-κB (96), members of the Raf/MEK/ERK pathway (36), and RTKs (121, 129), are known to facilitate IAV replication. Among the thousands of virus-supportive host factors identified, only a relatively small fraction show overlap in multiple independent screens (e.g., 113, 14, and 6 factors were found to be common in two, three, and four individual screens, respectively). It is noteworthy that no common host factors have been identified among all seven IAV RNAi screens performed to date. We therefore can conclude that RNAi screens can provide a holistic view on host dependency factors for virus replication. However, data reproducibility among different screens (due to various inherent or other uncontrollable variability issues) and determination of definitive mechanistic insights for the involvement of many of the different host factors in virus replication can be challenging (356). This problem can perhaps be overcome by the use of a lentivirus-based pooled RNAi screen. In this method, cells are first infected with a pool of lentiviruses at a low multiplicity of infection (MOI) (0.1 to 0.3) to express the diverse pool of siRNAs. Subsequently, cells are challenged with the target virus of interest in order to produce cytopathic effects. The surviving cells are then propagated and used for next-generation sequencing (NGS). This identifies siRNA targets (host genes) responsible for providing protection against virus-induced cell death (329, 330).
CRISPR/Cas9 screens.
As discussed above, the RNAi approach to identify exploitable host targets for antiviral drug development can be challenging, but the recently developed CRISPR/Cas9 approach portends to be an improved method. While RNAi produces a weak phenotype (partly because it is not possible to achieve 100% transfection efficiency), the cellular CRISPR/Cas9 machinery completely disrupts its targeted protein and thereby can produce a more robust phenotype (357). The genome-wide CRISPR/Cas9 knockout (GeCKO) technique can successfully identify cellular factors required for virus replication (358–367). This approach has many advantages over RNAi screens. For example, while the number of proviral host factors identified by seven IAV RNAi screens ranged from 90 to 323, the GeCKO screen identified a maximum of 453 proinfluenza host genes (359). Furthermore, the GeCKO screen identified at least 33 common proinfluenza host genes, compared to 2 to 16 identified by various RNAi screens (359). The GeCKO screen also revealed >400 rare host genes required for IAV replication that were not found in previous RNAi screens (359). Likewise, three independent GeCKO screens with flaviviruses (FLVs) identified the endoplasmic reticulum (ER)-associated protein complex as key cellular factors for efficient FLV replication. This indicated a higher reproducibility than that of RNAi screens (360, 362, 363). HIV-1 coreceptors, namely, solute carrier family 35 member B2 (SLC35B2), activated leukocyte cell adhesion molecule (ALCAM), and tyrosylprotein sulfotransferase 2 (TPST2) (336–341), as well as norovirus (NV) cellular receptor CD300If (367), were also identified by GeCKO screens.
In loss-of-function-based GeCKO/RNAi screens, identification of host restriction or dependency factors is based on the increase or decrease in viral titers. However, certain cellular factors do not directly contribute in regulating virus production but instead regulate cell death pathways. For example, Ma and colleagues have identified EMC2, EMC3, SELL1, DERL2, UBE2G2, UBE2J1, and HRD1 as highly enriched genes, all of which belong to ER-associated protein degradation (ERAD) pathways in a GeCKO screen for WNV. These genes exhibit no impact on virus production but have been associated with WNV-induced cell death (364).
Several factors must be considered when conducting and interpreting data produced by RNAi and GeCKO screens. Considering the level of genetic redundancy in animals, the variable levels of the efficiency of RNAi and GeCKO screens, and the diversity of experimental conditions, off-target effects that contribute to the relatively high rate of false-positive results can sometimes occur. This coupled with the relatively low rate of verification of the selected genes highlights the need for validation to confirm that the identified host genes are indeed coopted for virus replication.
CHALLENGES IN DEVELOPING HOST-DIRECTED THERAPIES
Drug Resistance against Host-Directed Antiviral Agents
As discussed before, since viruses cannot easily replace the missing cellular functions by mutagenesis, it is thought that viruses are unlikely to develop drug resistance against agents that target host factors needed for virus replication (37). However, emerging evidences suggest that viral resistance against host-directed antiviral agents can indeed occur (Table 4). In a cell culture model, clinically relevant IAV-directed agents may induce a completely resistant phenotype in a short period of time, such as after six passages (P6) (129). However, at this passage level, no drug-resistant IAV variants are known to be selected in the presence of host-directed agents in cell culture (129). Other attempts have also failed to derive drug-resistant virus variants against host-directed agents at up to ∼P25 (129, 368, 369). However, upon further passaging of virus in the presence of host-directed agents in cell culture (>P35), in many instances, viral substrains with partial but highly significant resistance phenotypes may be observed (39). Further virus propagation (up to P70) in the presence of host-directed agents did not appear to increase the magnitude of drug resistance (39). Interestingly, drug-resistant viruses maintained their phenotype upon withdrawal of the inhibitor from the cell culture medium (our unpublished data). These lines of evidence have demonstrated that resistant virus variants against host-directed agents cannot be easily generated but may still occur at a relatively low level upon long-term exposure to the host-directed agents. In this section, we will discuss the potential mechanisms that may underlie the emergence of drug resistance against host-directed antiviral agents.
TABLE 4.
Observations on development of resistance against host-directed antiviral agentsa
| Antiviral agent(s) | Virus | Host target | Mechanism of resistance | Reference(s) |
|---|---|---|---|---|
| Thapsigargin | NDV | SERCA | Mutations in the fusion (F) protein | 39 |
| Amiloride (EIPA) | CVB3 | Sodium-proton exchange | Mutations close to the active center in the RNA-dependent RNA polymerase | 430 |
| CGP57380 | BPXV | MNK1 | Not defined | 38 |
| DEBIO-025, SCY63, NIM811 | HCV | Cyclophilin | Mutations in the viral proteins (NS3, NS5A, and NS5B) near cyclophilin binding site | 372 |
| PIK93, BF738735, GW5074, T-00127-HEV1 | EV | PI4KB | Mutation in viral 3A protein which allows recruitment of PI4KB for synthesis of PI4P with enhanced efficiency | 481, 482 |
| Brequinar | DENV | Not defined | Mutations in the viral polymerase (E802Q) and envelope (M260V) proteins | 525 |
Abbreviations: BPXV, buffalopox virus; CVB3, coxsackievirus B3; DENV, dengue virus; EV, enterovirus; HCV, hepatitis C virus; NDV, Newcastle disease virus; SERCA, sarco/endoplasmic reticulum calcium-ATPase; MNK1, MAPK interacting kinase 1; PI4KB, phosphatidylinositol 4-kinase IIIβ.
Switch to use alternate host factor(s).
While not yet fully understood, one possible mechanism underlying the acquisition of drug resistance against host-directed agents is that the virus may switch to use an alternate host factor(s) (370) (Fig. 3A). For instance, propagation of HCV in CLDN1 knockout cells can generate CLDN1-independent HCV variants which can successfully infect and replicate in cells by using alternative host proteins—CLDN6 or CLDN9 (370).
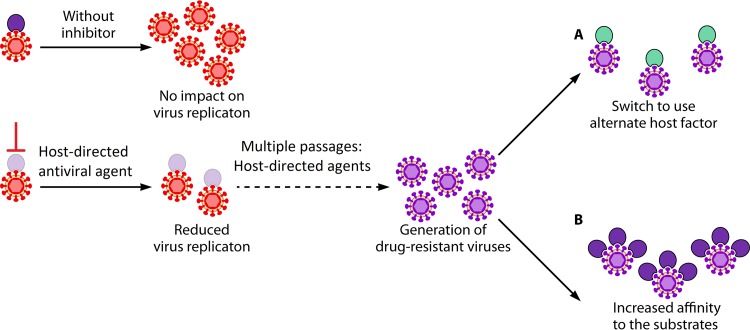
FIG 3.
Potential mechanisms underlying acquisition of resistance against host-directed antiviral agents. Three possible mechanisms that may be associated with acquisition of resistance against host-directed antiviral agents have been hypothesized. (A) Switch to use alternate host factor. Long-term restricted availability of a critical cellular factor may induce the virus to use an alternate host factor to become resistant. (B) Increased affinity to substrates. If viruses are cultured long term in the presence of restricted availability of a particular host factor, they may increase their efficiency to optimally replicate under a limiting amount of the targeted host protein.
Other examples include viruses that have evolved diverse strategies to modulate host translational apparatus. For example, many RNA viruses can disrupt (inactivate) cellular eIF4F to shut down initiation of cap-dependent translation of cellular proteins (37). However, they initiate their own mRNA translation via a cap-independent process that involves the internal ribosome entry site (IRES). In most DNA viruses and in a few RNA viruses, viral translation involves a cap-dependent mechanism, and this is mediated via the ERK/MNK1/eIF4E pathway (37). Activated MNK1 interacts with elF4G in the initiation complex and phosphorylates elF4E, which eventually binds to the 5′ cap of the mRNA to initiate translation (37). Prolonged passaging of BPXV in cell culture in the presence of chemical inhibitors targeting MNK1 or eIF4E has resulted in the generation of mutant viruses that are resistant to MNK1 or eIF4E inhibitor (38). While the exact molecular mechanism(s) of resistance remains unknown, it is tempting to suggest that the resistant virus may have switched to use an alternate host factor(s). Alternatively, it is possible that BPXV may switch to use an alternate (cap-independent) pathway of translational initiation (371). More work needs to be done to examine these possibilities.
Another example involves SERCA, which is a key cellular factor to support Newcastle disease virus (NDV) entry as well as synthesis and the subcellular localization of viral proteins (39). NDV mutants that efficiently replicate in the presence of SERCA inhibitor (thapsigargin) have emerged at ∼P40 in cell culture, although completely drug-resistant phenotypes have not been observed (39). At least one drug resistance-associated mutation (E104K) in the F protein of the mutant virus has been identified. Additional studies on recombinant NDVs that carry a point mutation(s) in either the F gene and/or other viral proteins are necessary in order to precisely uncover the molecular mechanism(s) underlying acquisition of drug resistance against thapsigargin.
Enhanced viral efficiency under selective pressure.
If viruses are cultured long term in the presence of restricted availability of a particular host factor, they may increase their efficiency to optimally replicate under limiting amounts of the drug-targeted host proteins (Fig. 3B). For example, depletion of cellular cyclophilin, a protein essential for HCV replication, results in reduced virus replication. However, long-term viral passage generates drug-resistant HCV variants that can replicate efficiently in cyclophilin-depleted cells. The resistant HCVs appear to have acquired mutations in their NS3, NS5A, and NS5B proteins, which show their higher affinity toward cyclophilin. This allows the resistant HCV strains to replicate optimally under limiting cyclophilin concentrations (372).
Phosphatidylinositol-4 kinase III β (PI4Kβ) can be a host target for enterovirus drug development (373). However, viral resistance mutations (G5318A or A70T) in the poliovirus 3A protein results in efficient virus growth in culture in the presence of the PI4Kβ inhibitor. Those virus mutants induce increased basal levels of phosphatidylinositol 4-phosphate (PI4P) lipid, which permits efficient viral replication in cell cultures depleted of PI4P (374). Interestingly, PI4Kβ-resistant coxsackievirus B3 mutants (3A-H57Y) can replicate in the presence of the PI4Kβ inhibitor without restoring high PI4P levels in the cell (375). This indicates that some mutation(s) in the coxsackievirus 3A genome can confer the resistance phenotype independent of PI4Kβ activation or PI4P lipid concentration. Likewise, cyclosporine (CsA)-resistant HCV mutants have also been identified in cell culture in the presence of CsA inhibitors (376, 377).
Synchronization of viral life cycle with patterns of antiviral drug therapy.
Some bacteriophages have evolved in such a way that the length of their life cycle is a mutable trait (378–383). Similarly, in bacteria, antibiotic tolerance (instead of resistance) is a process of temporarily surviving under high drug concentrations (384). The tolerance phenotype can result from mutations and may be heritable. It may involve changes in the timing of various steps of the life cycle of the organism (385, 386).
The most commonly understood mechanism of antiviral drug resistance against virus-directed therapies is that mutations occur in the viral genome at druggable sites and that these alter viral susceptibility to the direct action of drugs. However, in 2000, Wahl and Nowak proposed the term “cryptic resistance,” which defines virus populations that have become resistant without acquiring mutations at the druggable sites (387). This hypothesis was based on the fact that the concentration of antiviral drug in patients is not necessarily constant during treatment, because treatments are administered at timed intervals, with drugs being metabolized regularly. Between the administrations of two doses, the concentration of the drug may diminish to noninhibitory levels. This “cryptic resistance” phenotype describes a situation where the virus may adapt its life cycle to replicate during the periods of lowest drug concentration so as to permit sustained viral replication. However, this notion has not been formally proven. Therefore, in order to differentiate between the ability of the virus to grow in sustained (drug-resistant) versus transiently (drug-tolerant) high drug concentrations, the “cryptic resistance” terminology has been modified to “drug tolerance by synchronization” (388). Using a mathematical model, Neagu and colleagues (388) showed adaptation of the viral life cycle in response to drug treatment, a process they have referred to as “synchronization of viral life cycle with patterns of antiviral drug therapy” as a mechanism of viral drug tolerance. This effect is feasible when the times of drug dosing and viral life cycle are closely matched (388). However, this idea is based on in silico mathematical modeling and therefore needs empirical evidence in the laboratory and/or clinical settings. Moreover, the precise nature of host factors that may regulate the phenomenon of drug tolerance remains elusive. In addition, model systems are required to evaluate drug resistance/synchronization under complex and dynamic settings, such as drug combinations, multiple viral infections (214), and seasonality.
Translation into In Vivo Settings
A point of contention is that host-directed therapies are artifacts of in vitro conditions but have little translational applications (389). For example, VX-497, an IMP dehydrogenase (IMPDH) inhibitor, can impair HCV replication in vitro but not in vivo (390–392). This might occur because the nature and availability of nucleotides may differ under in vitro and in vivo conditions, thereby affecting the antiviral potency of the IMPDH inhibitor (368). Another example involves statins, which can have potent anti-HCV activity in vitro (390, 391, 393–395) but no clinical efficacy (396–399). This might partly be explained by statin activity being blocked in vivo due to its interaction with cholesterol.
Since host-directed agents interfere with host cell metabolism, a greater risk of cytotoxicity may be expected. As mentioned previously, while PI4Kβ inhibitors are known to exert potent antiviral activity against enteroviruses, they may prove to be lethal in mice, preventing their further development as antiviral drugs (400). Treating acute viral infections encounters fewer problems than treating chronic infections when host-directed agents are used (401). This is because a large number of host-directed compounds have inherent cellular toxicity problems if they are used for an extended period of time against chronic viral diseases. That being said, the majority of host-directed drugs that are in clinical use against cardiovascular and inflammatory diseases or cancers have minimal or no adverse side effects (402). For example, erlotinib (epidermal growth factor receptor [EGFR] inhibitor), an FDA-approved drug for non-small-cell lung cancer, is safe and well tolerated in patients with lung cancer (403). Similarly, clinically approved host-directed HIV-1 entry inhibitors have exhibited no reported adverse reactions (404). Nevertheless, the potential safety issue of host-directed antiviral agents remains a major concern and needs to be critically analyzed.
CONCLUDING REMARKS
At the time when complete information about the human genome/kinome was not available, reductionist approaches could typically identify only a single antiviral host target at a time. The availability of libraries of small molecule chemical inhibitors against a wide range of kinases, phosphatases, and other host factors has made it possible to rapidly and simultaneously screen multiple cellular factors required for virus replication. In this review, we make the case for designing drugs that target some of these host factors, since it is unlikely for a virus to replace the functions of missing host factors by mutagenesis. However, there are examples where the virus can either switch to use an alternate host factor(s) or alter its affinity toward the normal host dependency factor(s). Our current understanding of virus replication and virus-host interactions is far from complete. Functional and mechanistic studies based on biochemical approaches (such as the use of cell extracts and single-molecule techniques) in combination with novel live-cell imaging technologies will be essential to unravel the roles of candidate cellular proteins in mediating virus replication. We need to add to the analysis various epigenetic processes that can be manipulated to control virus replication. Finally, we advocate that selecting antiviral agents that can interfere with multiple steps of the virus replication cycle along with combining agents that have the capability to restrict both virus growth and hypercytokinemia is a favorable strategy. Additionally, combining virus-directed agents and host-directed agents is a useful approach, as some combinations can have beneficial synergistic effects. However, developing efficient host-directed antiviral agents with low cytotoxicity and high tolerability that enhance patient compliance and drug administration may be a challenging task. Understanding the use and implications of precision (individualized) medicine, particularly in the setting of antiviral therapy, is comparatively new and will take time to adopt. In the future, as medical prescriptions and treatments are likely be more personalized, rapid and cost-effective technologies that can provide comprehensive information (“omics” data) of the individual patient will be essential.
ACKNOWLEDGMENTS
This work was supported in part by Science and Engineering Research Board (India) grant no. SB/SO/AS-20/2014 and CRG/004747/2018 to N. Kumar.
The funding agency had no role in design, data collection and interpretation, or the decision to submit this work for publication.
Biographies

Naveen Kumar, Ph.D., completed his B.V.Sc. (2000) and M.V.Sc. (2002) degrees from College of Veterinary Sciences, Bikaner, India, and his Ph.D. (2006) from the Friedrich Loeffler Institute, Insel Riems, Germany, and CCS Haryana Agricultural University, Hisar, India, under a sandwich program. He served as a postdoc at Emory University, Atlanta, Georgia, USA, from 2006 to 2011 and worked on understanding the interactions of influenza virus with host cell signaling pathways. He then joined the Indian Council of Agricultural Research in 2011 as a senior scientist and is currently serving as principal scientist at ICAR–National Centre for Veterinary Type Cultures, Hisar, India. He is an OIE-designated member of the research group on pestes des petits ruminants. Dr. Kumar’s research interest is in the area of infectious diseases of animals, with a focus on understanding virus-host interactions for antiviral drug development.

Shalini Sharma, Ph.D., completed her B.V.Sc. (2002) and M.V.Sc. (2004) degrees from College of Veterinary Sciences, Bikaner, India, and her Ph.D. from University of Tennessee, Knoxville, Tennessee, USA (2011). She worked on understanding immunity and immunopathology to acute viral infections during her doctoral research work. Thereafter, she served as a postdoc from 2011 to 2014 at St. Jude Children’s Research Hospital, Memphis, Tennessee, USA, where she worked on immunity and immunopathology to heterologous viral infections. Since 2014, she has been working as an assistant professor at Lala Lajpat Rai University of Veterinary and Animal Sciences, Hisar, Haryana, India. Her area of interest is infectious diseases, with a focus on understanding cellular immune response against selected pathogens of domestic livestock.

Ram Kumar, M.V.Sc., completed his B.V.Sc. (2016) and M.V.Sc. (2018) degrees from Rajasthan University of Veterinary and Animal Science, Bikaner, India. During his master’s degree research work, he explored the role of MNK1 in initiation of buffalopox virus protein translation. Currently, he is pursuing his Ph.D. research work at the National Centre for Veterinary Type Cultures, Hisar, India, and working on the epigenetic regulation of paramyxovirus replication.

Bhupendra N. Tripathi, Ph.D., obtained his B.V.Sc. & A.H. degree from CSA University of Agriculture & Technology, Kanpur, India, and his M.V.Sc (1987) and Ph.D. (1990) degrees from the Indian Veterinary Research Institute (IVRI), Izatnagar, India. He joined IVRI in 1993 and worked there in the capacity of scientist, senior scientist, and principal scientist before moving to CSWRI, Avikanagar, India (2009), as the head of the Animal Health Division. He also served as a postdoc fellow at the Institute of Animal Health Compton, United Kingdom, and at Moredun Research Institute, Edinburgh, United Kingdom. Currently, he is the director at ICAR–National Research Centre on Equines and the project coordinator of the National Centre for Veterinary Type Cultures. Dr. Tripathi is a veterinary pathologist and has contributed significantly to understanding the pathogenesis of PPR and influenza viruses, including molecular virology to in vivo pathogenesis.

Sanjay Barua, Ph.D., completed his B.V.Sc. degree from the College of Veterinary Science, Guwahati, India, in 1991 and his M.V.Sc. degree (Veterinary Virology) from the Indian Veterinary Research Institute (IVRI), Izatnagar, India, in 1994. He joined the Central Institute for Research on Goats, Mathura, India, as scientist (veterinary microbiology) in 1997. He was awarded his Ph.D. degree in 2002 by IVRI, Izatnagar, India. He joined the National Centre for Veterinary Type Cultures (NCVTC), Hisar, India, in 2006 and is presently serving as the head of the national repository of animal microbes. He is particularly interested in understanding the role of host cell kinases in virus replication, in addition to exploring the diversity of the viruses in domestic animals across India.

Hinh Ly, Ph.D., received his B.S. and M.A. degrees with honors in microbiology and molecular genetics from the University of California, Los Angeles (UCLA), in 1995 and 1998, respectively, and his Ph.D. degree in microbiology and immunology from the University of North Carolina at Chapel Hill (UNC) in 2000. He carried out his postdoctoral work at the University of California, San Francisco (UCSF) (2000-2003), in the laboratories of Dr. Tristram Parslow (HIV researcher) and Dr. Elizabeth Blackburn (telomere researcher and 2009 Nobel Prize winner in physiology or medicine). Prior to his current position as a professor in the Department of Veterinary Biomedical Sciences at the University of Minnesota, Twin Cities, Dr. Ly served as an assistant professor in the Department of Pathology and Laboratory Medicine at Emory University in Atlanta. Dr. Ly's research interests have focused on virus-host interactions and vaccine and antiviral developments. Work in his laboratory has been consistently supported by the National Institutes of Health (NIH), the Department of Agriculture-National Institute of Food and Agriculture (USDA-NIFA), and the Minnesota Agricultural Experiment Station. He serves as a frequent reviewer of grant applications for the NIH, the Department of Defense (DoD), and other national and international agencies. He is currently serving on the editorial board of several scientific journals (e.g., Journal of Virology, Vaccines, Pathogens, and Virulence) and is Deputy Editor-in-Chief of the Journal of Medical Virology.

Barry T. Rouse, Ph.D., obtained his B.V.Sc degree from University of Bristol, England (1965), and his M.Sc. (1967) and Ph.D. (1970) degrees from the University of Guelph, Canada. He worked as a postdoc fellow at the Walter and Eliza Hall Institute of Medical Research, Australia (1970-72), and also earned his D.Sc. from University of Bristol, England (1997). Currently, he is a distinguished professor at the Department of Biomedical and Diagnostic Sciences, University of Knoxville, Tennessee, USA. He has been extensively involved in reviewing NIH grants since 1978 and has been a member of Faculty of 1000 since its inception. Dr. Rouse’s research interest is in the field of infectious disease and has focused on viral immunology and immunopathology. Dr. Rouse’s group was the first to show the role of regulatory T cells (Treg) in the host response to a virus infection.
REFERENCES
- 1.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Davison AJ. 2019. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol 164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- 2.Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R. 2020. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19). Int J Surg 76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ling L, Joynt GM, Lipman J, Constantin JM, Joannes-Boyau O. 20 February 2020. COVID-19: a critical care perspective informed by lessons learnt from other viral epidemics. Anaesth Crit Care Pain Med doi: 10.1016/j.accpm.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetter P, Eckerle I, Kaiser L. 2020. Covid-19: a puzzle with many missing pieces. BMJ 368:m627. doi: 10.1136/bmj.m627. [DOI] [PubMed] [Google Scholar]
- 5.Heymann DL, Shindo N, WHO Scientific Technical Advisory Group for Infectious Hazards . 2020. COVID-19: what is next for public health? Lancet 395:542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross AG, Olveda RM, Yuesheng L. 2014. Are we ready for a global pandemic of Ebola virus? Int J Infect Dis 28:217–218. doi: 10.1016/j.ijid.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Stech J, Beer M, Vahlenkamp T, Harder T. 2010. The pandemic influenza virus H1N1/2009: a review of the molecular biology, phylogeny, history of reassortments, and parameters of host switching. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 53:1231–1237. (In German.) doi: 10.1007/s00103-010-1166-0. [DOI] [PubMed] [Google Scholar]
- 8.Ligon BL. 2005. Avian influenza virus H5N1: a review of its history and information regarding its potential to cause the next pandemic. Semin Pediatr Infect Dis 16:326–335. doi: 10.1053/j.spid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Rolleston JD. 1933. The smallpox pandemic of 1870-1874: (Section of Epidemiology and State Medicine). Proc R Soc Med 27:177–192. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu ZB, Zhong CK, Zhang KX, Dong C, Peng H, Xu T, Wang AL, Guo ZR, Zhang YH. 2020. Epidemic trend of corona virus disease 2019 (COVID-19) in mainland China. Zhonghua Yu Fang Yi Xue Za Zhi 54:E022 (In Chinese.) doi: 10.3760/cma.j.cn112150-20200222-00163. [DOI] [PubMed] [Google Scholar]
- 11.Nellore A, Fishman J. 2009. Pandemic swine flu 2009. Xenotransplantation 16:463–465. doi: 10.1111/j.1399-3089.2009.00559.x. [DOI] [PubMed] [Google Scholar]
- 12.Rioux Y. 2006. Influenza, bird flu and pandemic. Perspect Infirm 4:29–33. (In French.) [PubMed] [Google Scholar]
- 13.Manderscheid RW. 2006. Preparing for a bird flu pandemic. Behav Healthc 26:43–44. [PubMed] [Google Scholar]
- 14.Flecknoe D, Charles Wakefield B, Simmons A. 2018. Plagues & wars: the 'Spanish Flu' pandemic as a lesson from history. Med Confl Surviv 34:61–68. doi: 10.1080/13623699.2018.1472892. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton D. 1992. Unanswered questions of the Spanish flu pandemic. Bull Am Assoc Hist Nurs 34:6–7. [PubMed] [Google Scholar]
- 16.Goris N, Vandenbussche F, De Clercq K. 2008. Potential of antiviral therapy and prophylaxis for controlling RNA viral infections of livestock. Antiviral Res 78:170–178. doi: 10.1016/j.antiviral.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 17.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingiliz P, Wehmeyer MH, Boesecke C, Schulze Zur Wiesch J, Schewe K, Lutz T, Baumgarten A, Simon KG, Hueppe D, Rockstroh JK, Mauss S, Christensen S, NEAT Study Group, GECCO Study Group . 28 September 2019. Reinfection with the hepatitis C virus in men who have sex with men after successful treatment with direct-acting antivirals in Germany: current incidence rates compared with rates during the interferon era. Clin Infect Dis doi: 10.1093/cid/ciz949. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham HE, Shea TC, Grgic T, Lachiewicz AM. 2019. Successful treatment of hepatitis C virus infection with direct-acting antivirals during hematopoietic cell transplant. Transpl Infect Dis 21:e13091. doi: 10.1111/tid.13091. [DOI] [PubMed] [Google Scholar]
- 20.Nagao Y, Kimura K, Kawahigashi Y, Sata M. 2016. Successful treatment of hepatitis C virus-associated oral lichen planus by interferon-free therapy with direct-acting antivirals. Clin Transl Gastroenterol 7:e179. doi: 10.1038/ctg.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri S, Symons JA, Deval J. 2018. Innovation and trends in the development and approval of antiviral medicines: 1987-2017 and beyond. Antiviral Res 155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li DK, Chung RT. 2019. Overview of direct-acting antiviral drugs and drug resistance of hepatitis C virus. Methods Mol Biol 1911:3–32. doi: 10.1007/978-1-4939-8976-8_1. [DOI] [PubMed] [Google Scholar]
- 23.Irwin KK, Renzette N, Kowalik TF, Jensen JD. 2016. Antiviral drug resistance as an adaptive process. Virus Evol 2:vew014. doi: 10.1093/ve/vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locarnini S, Bowden S. 2010. Drug resistance in antiviral therapy. Clin Liver Dis 14:439–459. doi: 10.1016/j.cld.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Hovi T, Jarvinen A, Pyhala R, Ristola M, Salminen M. 2002. Viruses and antiviral drug resistance. Duodecim 118:911–918. (In Finnish.) [PubMed] [Google Scholar]
- 26.Pillay D, Zambon M. 1998. Antiviral drug resistance. BMJ 317:660–662. doi: 10.1136/bmj.317.7159.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oxford JS. 1976. Drug resistance and antiviral agents. J Antimicrob Chemother 2:223–224. doi: 10.1093/jac/2.3.223. [DOI] [PubMed] [Google Scholar]
- 28.Venter JC, Smith HO, Adams MD. 2015. The sequence of the human genome. Clin Chem 61:1207–1208. doi: 10.1373/clinchem.2014.237016. [DOI] [PubMed] [Google Scholar]
- 29.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, et al. . 2001. The sequence of the human genome. Science 291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 30.McDougall WM, Perreira JM, Reynolds EC, Brass AL. 2018. CRISPR genetic screens to discover host-virus interactions. Curr Opin Virol 29:87–100. doi: 10.1016/j.coviro.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Gebre M, Nomburg JL, Gewurz BE. 2018. CRISPR-Cas9 genetic analysis of virus-host interactions. Viruses 10:55. doi: 10.3390/v10020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puschnik AS, Majzoub K, Ooi YS, Carette JE. 2017. A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol 15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halehalli RR, Nagarajaram HA. 2015. Molecular principles of human virus protein-protein interactions. Bioinformatics 31:1025–1033. doi: 10.1093/bioinformatics/btu763. [DOI] [PubMed] [Google Scholar]
- 34.Brito AF, Pinney JW. 2017. Protein-protein interactions in virus-host systems. Front Microbiol 8:1557. doi: 10.3389/fmicb.2017.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Wakker SI, Fischer MJE, Oosting RS. 2017. New drug-strategies to tackle viral-host interactions for the treatment of influenza virus infections. Eur J Pharmacol 809:178–190. doi: 10.1016/j.ejphar.2017.05.038. [DOI] [PubMed] [Google Scholar]
- 36.Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol 3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 37.Kumar R, Khandelwal N, Thachamvally R, Tripathi BN, Barua S, Kashyap SK, Maherchandani S, Kumar N. 2018. Role of MAPK/MNK1 signaling in virus replication. Virus Res 253:48–61. doi: 10.1016/j.virusres.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar R, Khandelwal N, Chander Y, Riyesh T, Tripathi BN, Kashyap SK, Barua S, Maherchandani S, Kumar N. 2018. MNK1 inhibitor as an antiviral agent suppresses buffalopox virus protein synthesis. Antiviral Res 160:126–136. doi: 10.1016/j.antiviral.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 39.Kumar N, Khandelwal N, Kumar R, Chander Y, Rawat KD, Chaubey KK, Sharma S, Singh SV, Riyesh T, Tripathi BN, Barua S. 2019. Inhibitor of sarco/endoplasmic reticulum calcium-ATPase impairs multiple steps of paramyxovirus replication. Front Microbiol 10:209. doi: 10.3389/fmicb.2019.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen AL. 2018. Host factors involved in Ebola virus replication. Curr Top Microbiol Immunol 419:113–150. doi: 10.1007/82_2017_27. [DOI] [PubMed] [Google Scholar]
- 41.Delpeut S, Noyce RS, Siu RW, Richardson CD. 2012. Host factors and measles virus replication. Curr Opin Virol 2:773–783. doi: 10.1016/j.coviro.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Mehle A, Doudna JA. 2010. A host of factors regulating influenza virus replication. Viruses 2:566–573. doi: 10.3390/v2020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Diedrichs-Mohring M, Wildner G. 2019. The role of IFN-alpha in experimental and clinical uveitis. Ocul Immunol Inflamm 27:23–33. doi: 10.1080/09273948.2017.1298822. [DOI] [PubMed] [Google Scholar]
- 44.Friedman RM, Contente S. 2010. Treatment of hepatitis C infections with interferon: a historical perspective. Hepat Res Treat 2010:323926. doi: 10.1155/2010/323926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo ASJ, Kwok R, Ahmed T. 2017. Alpha-interferon treatment in hepatitis B. Ann Transl Med 5:159. doi: 10.21037/atm.2017.03.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beglin M, Melar-New M, Laimins L. 2009. Human papillomaviruses and the interferon response. J Interferon Cytokine Res 29:629–635. doi: 10.1089/jir.2009.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedder SC. 2003. Pegylation of interferon alfa: structural and pharmacokinetic properties. Semin Liver Dis 23(Suppl 1):19–22. [DOI] [PubMed] [Google Scholar]
- 48.Maughan A, Ogbuagu O. 2018. Pegylated interferon alpha 2a for the treatment of hepatitis C virus infection. Expert Opin Drug Metab Toxicol 14:219–227. doi: 10.1080/17425255.2018.1421173. [DOI] [PubMed] [Google Scholar]
- 49.Tsuge M, Uchida T, Hiraga N, Kan H, Makokha GN, Abe-Chayama H, Miki D, Imamura M, Ochi H, Hayes CN, Shimozono R, Iwamura T, Narumi H, Suzuki T, Kainoh M, Taniguchi T, Chayama K. 2017. Development of a novel site-specific pegylated interferon beta for antiviral therapy of chronic hepatitis B virus. Antimicrob Agents Chemother 61:e00183-17. doi: 10.1128/AAC.00183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osinusi A, Rasimas JJ, Bishop R, Proschan M, McLaughlin M, Murphy A, Cortez KJ, Polis MA, Masur H, Rosenstein D, Kottilil S. 2010. HIV/hepatitis C virus-coinfected virologic responders to pegylated interferon and ribavirin therapy more frequently incur interferon-related adverse events than nonresponders do. J Acquir Immune Defic Syndr 53:357–363. doi: 10.1097/QAI.0b013e3181c7a29d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuda F, Torii Y, Enomoto H, Kuga C, Aizawa N, Iwata Y, Saito M, Imanishi H, Shimomura S, Nakamura H, Tanaka H, Iijima H, Tsutsui H, Tanaka Y, Nishiguchi S. 2012. Anti-interferon-alpha neutralizing antibody is associated with nonresponse to pegylated interferon-alpha plus ribavirin in chronic hepatitis C. J Viral Hepat 19:694–703. doi: 10.1111/j.1365-2893.2012.01598.x. [DOI] [PubMed] [Google Scholar]
- 52.Tsubota A, Fujise K, Namiki Y, Tada N. 2011. Peginterferon and ribavirin treatment for hepatitis C virus infection. World J Gastroenterol 17:419–432. doi: 10.3748/wjg.v17.i4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung RT, Gale M Jr, Polyak SJ, Lemon SM, Liang TJ, Hoofnagle JH. 2008. Mechanisms of action of interferon and ribavirin in chronic hepatitis C: summary of a workshop. Hepatology 47:306–320. doi: 10.1002/hep.22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tai AW, Chung RT. 2009. Treatment failure in hepatitis C: mechanisms of non-response. J Hepatol 50:412–420. doi: 10.1016/j.jhep.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, Heathcote EJ, Zeuzem S, Reesink HW, Garg J, Bsharat M, George S, Kauffman RS, Adda N, Di Bisceglie AM, PROVE3 Study Team . 2010. Telaprevir for previously treated chronic HCV infection. N Engl J Med 362:1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 56.Lang L. 2007. Combination therapy with telaprevir and pegylated interferon suppresses both wild-type and resistant hepatitis C virus. Gastroenterology 132:5–6. doi: 10.1053/j.gastro.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 57.Laguno M, Von Wichmann MA, Van den Eynde E, Navarro J, Cifuentes C, Murillas J, Veloso S, Martinez-Rebollar M, Guardiola JM, Jou A, Gomez-Sirvent JL, Cervantes M, Pineda JA, Lopez-Calvo S, Carrero A, Montes ML, Deig E, Tapiz A, Ruiz-Mesa JD, Cruceta A, de Lazzari E, Mallolas J. 2016. Boceprevir plus pegylated interferon/ribavirin to re-treat hepatitis C virus genotype 1 in HIV-HCV co-infected patients: final results of the Spanish BOC HIV-HCV Study. Int J Infect Dis 53:46–51. doi: 10.1016/j.ijid.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 58.Tamai H, Ida Y, Kawashima A, Shingaki N, Shimizu R, Moribata K, Nasu T, Maekita T, Iguchi M, Kato J, Nakao T, Kitano M. 2017. Simeprevir-based triple therapy with reduced doses of pegylated interferon alpha-2a plus ribavirin for interferon ineligible patients with genotype 1b hepatitis C virus. Gut Liver 11:551–558. doi: 10.5009/gnl16525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mucenic M, Brandao ABM, Marroni CA, Fleck Junior AM, Zanotelli ML, Leipnitz I, Meine MH, Kiss G, Martini J, Schlindwein ES, Costabeber AM, Sacco FKR, Rossato G, Cantisani GPC. 2019. Sofosbuvir, ribavirin and pegylated interferon for a daclatasvir-resistant genotype 3 hepatitis C virus: case report and review. Rev Inst Med Trop Sao Paulo 61:e12. doi: 10.1590/S1678-9946201961012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Son BK, Sohn JH, Kim TY, Park YK, Jeon YC, Han DS. 2007. Pulmonary toxicity by pegylated interferon alpha-2a in a patient with chronic hepatitis C. Korean J Hepatol 13:103–107. (In Korean.) [PubMed] [Google Scholar]
- 61.Chisholm JA, Williams G, Spence E, Parks S, Keating D, Gavin M, Mills PR. 2005. Retinal toxicity during pegylated alpha-interferon therapy for chronic hepatitis C: a multifocal electroretinogram investigation. Aliment Pharmacol Ther 21:723–732. doi: 10.1111/j.1365-2036.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- 62.Shindo M, Terai I. 2013. Adverse skin reactions due to ribavirin in hepatitis C combination therapy with pegylated interferon-alpha2a. Case Rep Dermatol 5:379–381. doi: 10.1159/000357516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hashimoto Y, Kanto H, Itoh M. 2007. Adverse skin reactions due to pegylated interferon alpha 2b plus ribavirin combination therapy in a patient with chronic hepatitis C virus. J Dermatol 34:577–582. doi: 10.1111/j.1346-8138.2007.00336.x. [DOI] [PubMed] [Google Scholar]
- 64.Saito H, Tada S, Ebinuma H, Ishii H, Kashiwazaki K, Takahashi M, Tsukada N, Nishida J, Tanaka S, Shiozaki H, Hibi T. 2006. Role of erythrocytes as a reservoir for ribavirin and relationship with adverse reactions in the early phase of interferon combination therapy for chronic hepatitis C virus infections. J Clin Microbiol 44:3562–3568. doi: 10.1128/JCM.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manjon-Haces JA, Vazquez-Lopez F, Gomez-Diez S, Hidalgo-Garcia A, Perez-Alvarez R, Soler-Sanchez T, Perez-Oliva N. 2001. Adverse cutaneous reactions to interferon alfa-2b plus ribavirin therapy in patients with chronic hepatitis C virus. Acta Derm Venereol 81:223. [DOI] [PubMed] [Google Scholar]
- 66.Vacchelli E, Galluzzi L, Eggermont A, Galon J, Tartour E, Zitvogel L, Kroemer G. 2012. Trial Watch: immunostimulatory cytokines. Oncoimmunology 1:493–506. doi: 10.4161/onci.20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gull I, Aslam MS, Tipu I, Mushtaq R, Ali TZ, Athar MA. 2019. Development of latent interferon alpha 2b as a safe therapeutic for treatment of hepatitis C virus infection. Sci Rep 9:10867. doi: 10.1038/s41598-019-47074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaudieri S, Rauch A, Pfafferott K, Barnes E, Cheng W, McCaughan G, Shackel N, Jeffrey GP, Mollison L, Baker R, Furrer H, Gunthard HF, Freitas E, Humphreys I, Klenerman P, Mallal S, James I, Roberts S, Nolan D, Lucas M. 2009. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49:1069–1082. doi: 10.1002/hep.22773. [DOI] [PubMed] [Google Scholar]
- 69.Pawlotsky JM. 2009. Therapeutic implications of hepatitis C virus resistance to antiviral drugs. Therap Adv Gastroenterol 2:205–219. doi: 10.1177/1756283X09336045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Serre SB, Krarup HB, Bukh J, Gottwein JM. 2013. Identification of alpha interferon-induced envelope mutations of hepatitis C virus in vitro associated with increased viral fitness and interferon resistance. J Virol 87:12776–12793. doi: 10.1128/JVI.00901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gale M Jr, Katze MG. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther 78:29–46. doi: 10.1016/S0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 72.van Broekhoven CL, Parish CR, Demangel C, Britton WJ, Altin JG. 2004. Targeting dendritic cells with antigen-containing liposomes: a highly effective procedure for induction of antitumor immunity and for tumor immunotherapy. Cancer Res 64:4357–4365. doi: 10.1158/0008-5472.CAN-04-0138. [DOI] [PubMed] [Google Scholar]
- 73.Pardridge WM. 2010. Biopharmaceutical drug targeting to the brain. J Drug Target 18:157–167. doi: 10.3109/10611860903548354. [DOI] [PubMed] [Google Scholar]
- 74.Coulstock E, Sosabowski J, Ovecka M, Prince R, Goodall L, Mudd C, Sepp A, Davies M, Foster J, Burnet J, Dunlevy G, Walker A. 2013. Liver-targeting of interferon-alpha with tissue-specific domain antibodies. PLoS One 8:e57263. doi: 10.1371/journal.pone.0057263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai G, Jiang M, Zhou Y, Gu X, Zhang B, Zou M, Zhou X, Bao J, Cao G, Zhang R. 2011. Generation of a liver targeting fusion interferon and its bioactivity analysis in vitro. Pharmazie 66:761–765. [PubMed] [Google Scholar]
- 76.Welander CE, Homesley HD, Smiles KA, Peets EA. 1990. Intralesional interferon alfa-2b for the treatment of genital warts. Am J Obstet Gynecol 162:348–354. doi: 10.1016/0002-9378(90)90383-i. [DOI] [PubMed] [Google Scholar]
- 77.Monk BJ, Tewari KS. 2007. The spectrum and clinical sequelae of human papillomavirus infection. Gynecol Oncol 107:S6–S13. doi: 10.1016/j.ygyno.2007.07.076. [DOI] [PubMed] [Google Scholar]
- 78.Slade HB, Owens ML, Tomai MA, Miller RL. 1998. Imiquimod 5% cream (Aldara). Expert Opin Investig Drugs 7:437–449. doi: 10.1517/13543784.7.3.437. [DOI] [PubMed] [Google Scholar]
- 79.Sen E, McLaughlin-Drubin M, Meyers C. 2005. Efficacy of two commercial preparations of interferon-alpha on human papillomavirus replication. Anticancer Res 25:1091–1100. [PubMed] [Google Scholar]
- 80.Roche M, Salimi H, Duncan R, Wilkinson BL, Chikere K, Moore MS, Webb NE, Zappi H, Sterjovski J, Flynn JK, Ellett A, Gray LR, Lee B, Jubb B, Westby M, Ramsland PA, Lewin SR, Payne RJ, Churchill MJ, Gorry PR. 2013. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology 10:43. doi: 10.1186/1742-4690-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lieberman-Blum SS, Fung HB, Bandres JC. 2008. Maraviroc: a CCR5-receptor antagonist for the treatment of HIV-1 infection. Clin Ther 30:1228–1250. doi: 10.1016/S0149-2918(08)80048-3. [DOI] [PubMed] [Google Scholar]
- 82.Lopez-Huertas MR, Jimenez-Tormo L, Madrid-Elena N, Gutierrez C, Rodriguez-Mora S, Coiras M, Alcami J, Moreno S. 2017. The CCR5-antagonist maraviroc reverses HIV-1 latency in vitro alone or in combination with the PKC-agonist Bryostatin-1. Sci Rep 7:2385. doi: 10.1038/s41598-017-02634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, Jenkins TM, Perros M, van der Ryst E. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol 80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gulick RM, Su Z, Flexner C, Hughes MD, Skolnik PR, Wilkin TJ, Gross R, Krambrink A, Coakley E, Greaves WL, Zolopa A, Reichman R, Godfrey C, Hirsch M, Kuritzkes DR, AIDS Clinical Trials Group 5211 Team . 2007. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-infected, treatment-experienced patients: AIDS clinical trials group 5211. J Infect Dis 196:304–312. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- 86.Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol 81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Landovitz RJ, Angel JB, Hoffmann C, Horst H, Opravil M, Long J, Greaves W, Fatkenheuer G. 2008. Phase II study of vicriviroc versus efavirenz (both with zidovudine/lamivudine) in treatment-naive subjects with HIV-1 infection. J Infect Dis 198:1113–1122. doi: 10.1086/592052. [DOI] [PubMed] [Google Scholar]
- 89.Zhou Y, Frey TK, Yang JJ. 2009. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium 46:1–17. doi: 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lorizate M, Krausslich HG. 2011. Role of lipids in virus replication. Cold Spring Harb Perspect Biol 3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stapleford KA, Miller DJ. 2010. Role of cellular lipids in positive-sense RNA virus replication complex assembly and function. Viruses 2:1055–1068. doi: 10.3390/v2051055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen H, Yamashita A, Nakakoshi M, Yokoe H, Sudo M, Kasai H, Tanaka T, Fujimoto Y, Ikeda M, Kato N, Sakamoto N, Shindo H, Maekawa S, Enomoto N, Tsubuki M, Moriishi K. 2013. Inhibitory effects of caffeic acid phenethyl ester derivatives on replication of hepatitis C virus. PLoS One 8:e82299. doi: 10.1371/journal.pone.0082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaur P, Munjhal A, Lal SK. 2011. Influenza virus and cell signaling pathways. Med Sci Monit 17:RA148–RA154. doi: 10.12659/msm.881801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kawai T, Akira S. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol 21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ludwig S, Pleschka S, Planz O, Wolff T. 2006. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell Microbiol 8:375–386. doi: 10.1111/j.1462-5822.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- 96.Kumar N, Xin ZT, Liang Y, Ly H, Liang Y. 2008. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J Virol 82:9880–9889. doi: 10.1128/JVI.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milanesi L, Petrillo M, Sepe L, Boccia A, D'Agostino N, Passamano M, Di Nardo S, Tasco G, Casadio R, Paolella G. 2005. Systematic analysis of human kinase genes: a large number of genes and alternative splicing events result in functional and structural diversity. BMC Bioinformatics 6(Suppl 4):S20. doi: 10.1186/1471-2105-6-S4-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu X, Yang Y, Zhao M, Bode L, Zhang L, Pan J, Lv L, Zhan Y, Liu S, Zhang L, Wang X, Huang R, Zhou J, Xie P. 2014. Proteomics reveal energy metabolism and mitogen-activated protein kinase signal transduction perturbation in human Borna disease virus Hu-H1-infected oligodendroglial cells. Neuroscience 268:284–296. doi: 10.1016/j.neuroscience.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen YF, Xia Y. 2019. Convergent perturbation of the human domain-resolved interactome by viruses and mutations inducing similar disease phenotypes. PLoS Comput Biol 15:e1006762. doi: 10.1371/journal.pcbi.1006762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nousiainen L, Sillanpaa M, Jiang M, Thompson J, Taipale J, Julkunen I. 2013. Human kinome analysis reveals novel kinases contributing to virus infection and retinoic-acid inducible gene I-induced type I and type III IFN gene expression. Innate Immun 19:516–530. doi: 10.1177/1753425912473345. [DOI] [PubMed] [Google Scholar]
- 101.Briedis KM, Starr A, Bourne PE. 2008. Analysis of the human kinome using methods including fold recognition reveals two novel kinases. PLoS One 3:e1597. doi: 10.1371/journal.pone.0001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meineke R, Rimmelzwaan GF, Elbahesh H. 2019. Influenza virus infections and cellular kinases. Viruses 11:171. doi: 10.3390/v11020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duong-Ly KC, Peterson JR. 2013. The human kinome and kinase inhibition. Curr Protoc Pharmacol Chapter 2:Unit2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bekerman E, Neveu G, Shulla A, Brannan J, Pu SY, Wang S, Xiao F, Barouch-Bentov R, Bakken RR, Mateo R, Govero J, Nagamine CM, Diamond MS, De Jonghe S, Herdewijn P, Dye JM, Randall G, Einav S. 2017. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J Clin Invest 127:1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnsamuel J, Eriksson S, Oliveira M, Tjarks W. 2005. Docking simulation with a purine nucleoside specific homology model of deoxycytidine kinase, a target enzyme for anticancer and antiviral therapy. Bioorg Med Chem 13:4160–4167. doi: 10.1016/j.bmc.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 106.Krishnan P, Gullen EA, Lam W, Dutschman GE, Grill SP, Cheng YC. 2003. Novel role of 3-phosphoglycerate kinase, a glycolytic enzyme, in the activation of l-nucleoside analogs, a new class of anticancer and antiviral agents. J Biol Chem 278:36726–36732. doi: 10.1074/jbc.M307052200. [DOI] [PubMed] [Google Scholar]
- 107.Burkhard K, Shapiro P. 2010. Use of inhibitors in the study of MAP kinases. Methods Mol Biol 661:107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Uehling DE, Harris PA. 2015. Recent progress on MAP kinase pathway inhibitors. Bioorg Med Chem Lett 25:4047–4056. doi: 10.1016/j.bmcl.2015.07.093. [DOI] [PubMed] [Google Scholar]
- 109.Kim DH, Sim T. 2012. Novel small molecule Raf kinase inhibitors for targeted cancer therapeutics. Arch Pharm Res 35:605–615. doi: 10.1007/s12272-012-0403-5. [DOI] [PubMed] [Google Scholar]
- 110.Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA, Grinsted L, Zazulina V, Smith P, Smith I, Crino L. 2013. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 111.Matsuda Y, Fukumoto M. 2011. Sorafenib: complexities of Raf-dependent and Raf-independent signaling are now unveiled. Med Mol Morphol 44:183–189. doi: 10.1007/s00795-011-0558-z. [DOI] [PubMed] [Google Scholar]
- 112.Michelangeli F, East JM. 2011. A diversity of SERCA Ca2+ pump inhibitors. Biochem Soc Trans 39:789–797. doi: 10.1042/BST0390789. [DOI] [PubMed] [Google Scholar]
- 113.Levitzki A, Gazit A. 1995. Tyrosine kinase inhibition: an approach to drug development. Science 267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 114.Sweeney SE, Firestein GS. 2006. Mitogen activated protein kinase inhibitors: where are we now and where are we going? Ann Rheum Dis 65(Suppl 3):iii83–iii88. doi: 10.1136/ard.2006.058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bogoyevitch MA, Boehm I, Oakley A, Ketterman AJ, Barr RK. 2004. Targeting the JNK MAPK cascade for inhibition: basic science and therapeutic potential. Biochim Biophys Acta 1697:89–101. doi: 10.1016/j.bbapap.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 116.Gangwal RP, Bhadauriya A, Damre MV, Dhoke GV, Sangamwar AT. 2013. p38 Mitogen-activated protein kinase inhibitors: a review on pharmacophore mapping and QSAR studies. Curr Top Med Chem 13:1015–1035. doi: 10.2174/1568026611313090005. [DOI] [PubMed] [Google Scholar]
- 117.Genovese MC. 2009. Inhibition of p38: has the fat lady sung? Arthritis Rheum 60:317–320. doi: 10.1002/art.24264. [DOI] [PubMed] [Google Scholar]
- 118.Chahrour O, Cairns D, Omran Z. 2012. Small molecule kinase inhibitors as anti-cancer therapeutics. Mini Rev Med Chem 12:399–411. doi: 10.2174/138955712800493915. [DOI] [PubMed] [Google Scholar]
- 119.Giroux V, Dagorn JC, Iovanna JL. 2009. A review of kinases implicated in pancreatic cancer. Pancreatology 9:738–754. doi: 10.1159/000199435. [DOI] [PubMed] [Google Scholar]
- 120.Tsatsanis C, Spandidos DA. 2000. The role of oncogenic kinases in human cancer (Review). Int J Mol Med 5:583–590. doi: 10.3892/ijmm.5.6.583. [DOI] [PubMed] [Google Scholar]
- 121.Kumar N, Liang Y, Parslow TG, Liang Y. 2011. Receptor tyrosine kinase inhibitors block multiple steps of influenza A virus replication. J Virol 85:2818–2827. doi: 10.1128/JVI.01969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim YS, Kawai A. 1998. Studies on the antiviral mechanisms of protein kinase inhibitors K-252a and KT5926 against the replication of vesicular stomatitis virus. Biol Pharm Bull 21:498–505. doi: 10.1248/bpb.21.498. [DOI] [PubMed] [Google Scholar]
- 123.Kim YS, Sagara J, Kawai A. 1995. Studies on the antiviral activity of protein kinase inhibitors against the replication of vesicular stomatitis virus. Biol Pharm Bull 18:895–899. doi: 10.1248/bpb.18.895. [DOI] [PubMed] [Google Scholar]
- 124.Esther A, Iftach S, Esther P. 1994. Inhibition of Moloney murine leukemia virus replication by tyrphostins, tyrosine kinase inhibitors. FEBS Lett 341:99–103. doi: 10.1016/0014-5793(94)80248-3. [DOI] [PubMed] [Google Scholar]
- 125.Hayashi K. 1992. Effect of protein kinase C inhibitors with different action mechanisms on Epstein-Barr virus replication. Intervirology 33:217–224. doi: 10.1159/000150254. [DOI] [PubMed] [Google Scholar]
- 126.Perwitasari O, Yan X, O'Donnell J, Johnson S, Tripp RA. 2015. Repurposing kinase inhibitors as antiviral agents to control influenza A virus replication. Assay Drug Dev Technol 13:638–649. doi: 10.1089/adt.2015.0003.drrr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schor S, Einav S. 2018. Repurposing of kinase inhibitors as broad-spectrum antiviral drugs. DNA Cell Biol 37:63–69. doi: 10.1089/dna.2017.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schang LM. 2005. Discovery of the antiviral activities of pharmacologic cyclin-dependent kinase inhibitors: from basic to applied science. Expert Rev Anti Infect Ther 3:145–149. doi: 10.1586/14787210.3.2.145. [DOI] [PubMed] [Google Scholar]
- 129.Kumar N, Sharma NR, Ly H, Parslow TG, Liang Y. 2011. Receptor tyrosine kinase inhibitors that block replication of influenza A and other viruses. Antimicrob Agents Chemother 55:5553–5559. doi: 10.1128/AAC.00725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pombo JP, Sanyal S. 2018. Perturbation of intracellular cholesterol and fatty acid homeostasis during flavivirus infections. Front Immunol 9:1276. doi: 10.3389/fimmu.2018.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heaton NS, Randall G. 2011. Multifaceted roles for lipids in viral infection. Trends Microbiol 19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thaker SK, Ch'ng J, Christofk HR. 2019. Viral hijacking of cellular metabolism. BMC Biol 17:59. doi: 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Royle J, Donald CL, Merits A, Kohl A, Varjak M. 2017. Differential effects of lipid biosynthesis inhibitors on Zika and Semliki Forest viruses. Vet J 230:62–64. doi: 10.1016/j.tvjl.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Merino-Ramos T, Vázquez-Calvo Á, Casas J, Sobrino F, Saiz J-C, Martín-Acebes MA. 2016. Modification of the host cell lipid metabolism induced by hypolipidemic drugs targeting the acetyl coenzyme A carboxylase impairs West Nile virus replication. Antimicrob Agents Chemother 60:307–315. doi: 10.1128/AAC.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karlas A, Berre S, Couderc T, Varjak M, Braun P, Meyer M, Gangneux N, Karo-Astover L, Weege F, Raftery M, Schonrich G, Klemm U, Wurzlbauer A, Bracher F, Merits A, Meyer TF, Lecuit M. 2016. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat Commun 7:11320. doi: 10.1038/ncomms11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A 107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martín-Acebes MA, Vázquez-Calvo Á, Saiz J-C. 2016. Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses. Prog Lipid Res 64:123–137. doi: 10.1016/j.plipres.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 138.Wakil SJ, Abu-Elheiga LA. 2009. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res 50(Suppl):S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Villareal VA, Rodgers MA, Costello DA, Yang PL. 2015. Targeting host lipid synthesis and metabolism to inhibit dengue and hepatitis C viruses. Antiviral Res 124:110–121. doi: 10.1016/j.antiviral.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang CY, Wong WW, Tsai HC, Chen YH, Kuo BS, Lynn S, Blazkova J, Clarridge KE, Su HW, Lin CY, Tseng FC, Lai A, Yang FH, Lin CH, Tseng W, Lin HY, Finstad CL, Wong-Staal F, Hanson CV, Chun TW, Liao MJ. 2019. Effect of anti-CD4 antibody UB-421 on HIV-1 rebound after treatment interruption. N Engl J Med 380:1535–1545. doi: 10.1056/NEJMoa1802264. [DOI] [PubMed] [Google Scholar]
- 141.Emu B, Fessel J, Schrader S, Kumar P, Richmond G, Win S, Weinheimer S, Marsolais C, Lewis S. 2018. Phase 3 study of ibalizumab for multidrug-resistant HIV-1. N Engl J Med 379:645–654. doi: 10.1056/NEJMoa1711460. [DOI] [PubMed] [Google Scholar]
- 142.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 143.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 146.Yamashita M, Iida M, Tada M, Shirasago Y, Fukasawa M, Nagase S, Watari A, Ishii-Watabe A, Yagi K, Kondoh M. 2015. Discovery of anti-claudin-1 antibodies as candidate therapeutics against hepatitis C virus. J Pharmacol Exp Ther 353:112–118. doi: 10.1124/jpet.114.217653. [DOI] [PubMed] [Google Scholar]
- 147.Shimizu Y, Shirasago Y, Kondoh M, Suzuki T, Wakita T, Hanada K, Yagi K, Fukasawa M. 2018. Monoclonal antibodies against occludin completely prevented hepatitis C virus infection in a mouse model. J Virol 92:e02258-17. doi: 10.1128/JVI.02258-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Catanese MT, Graziani R, von Hahn T, Moreau M, Huby T, Paonessa G, Santini C, Luzzago A, Rice CM, Cortese R, Vitelli A, Nicosia A. 2007. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. J Virol 81:8063–8071. doi: 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Catanese MT, Loureiro J, Jones CT, Dorner M, von Hahn T, Rice CM. 2013. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol 87:8282–8293. doi: 10.1128/JVI.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, Pietschmann T. 2010. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog 6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck ZY, Foung SK, Pecheur EI, Pietschmann T. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol 84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vercauteren K, Van Den Eede N, Mesalam AA, Belouzard S, Catanese MT, Bankwitz D, Wong-Staal F, Cortese R, Dubuisson J, Rice CM, Pietschmann T, Leroux-Roels G, Nicosia A, Meuleman P. 2014. Successful anti-scavenger receptor class B type I (SR-BI) monoclonal antibody therapy in humanized mice after challenge with HCV variants with in vitro resistance to SR-BI-targeting agents. Hepatology 60:1508–1518. doi: 10.1002/hep.27196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Notario L, Redondo-Anton J, Alari-Pahissa E, Albentosa A, Leiva M, Lopez D, Sabio G, Lauzurica P. 2019. CD69 targeting enhances anti-vaccinia virus immunity. J Virol 93:e00553-19. doi: 10.1128/JVI.00553-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Donnelly RP, Loftus RM, Keating SE, Liou KT, Biron CA, Gardiner CM, Finlay DK. 2014. mTORC1-dependent metabolic reprogramming is a prerequisite for NK cell effector function. J Immunol 193:4477–4484. doi: 10.4049/jimmunol.1401558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cibrian D, Sanchez-Madrid F. 2017. CD69: from activation marker to metabolic gatekeeper. Eur J Immunol 47:946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sharma S, Mulik S, Kumar N, Suryawanshi A, Rouse BT. 2011. An anti-inflammatory role of VEGFR2/Src kinase inhibitor in herpes simplex virus 1-induced immunopathology. J Virol 85:5995–6007. doi: 10.1128/JVI.00034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hosseini H, Khalili MR. 2007. Therapeutic potential of bevacizumab (Avastin) in herpetic stromal keratitis (HSK). Med Hypotheses 69:568–570. doi: 10.1016/j.mehy.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 158.Halota W, Ferenci P, Kozielewicz D, Dybowska D, Lisovoder N, Samira S, Shalit I, Ellis R, Ilan Y. 2015. Oral anti-CD3 immunotherapy for HCV-nonresponders is safe, promotes regulatory T cells and decreases viral load and liver enzyme levels: results of a phase-2a placebo-controlled trial. J Viral Hepat 22:651–657. doi: 10.1111/jvh.12369. [DOI] [PubMed] [Google Scholar]
- 159.Zahid MN, Turek M, Xiao F, Thi VL, Guerin M, Fofana I, Bachellier P, Thompson J, Delang L, Neyts J, Bankwitz D, Pietschmann T, Dreux M, Cosset FL, Grunert F, Baumert TF, Zeisel MB. 2013. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology 57:492–504. doi: 10.1002/hep.26097. [DOI] [PubMed] [Google Scholar]
- 160.Lacek K, Vercauteren K, Grzyb K, Naddeo M, Verhoye L, Słowikowski MP, Fafi-Kremer S, Patel AH, Baumert TF, Folgori A, Leroux-Roels G, Cortese R, Meuleman P, Nicosia A. 2012. Novel human SR-BI antibodies prevent infection and dissemination of HCV in vitro and in humanized mice. J Hepatol 57:17–23. doi: 10.1016/j.jhep.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 161.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ, McKeating JA, Dragic T, Pessaux P, Stoll-Keller F, Schuster C, Thompson J, Baumert TF. 2010. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139:953–964.e4. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 162.Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, Desombere I, Reiser H, Leroux-Roels G. 2008. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 163.Fofana I, Xiao F, Thumann C, Turek M, Zona L, Tawar RG, Grunert F, Thompson J, Zeisel MB, Baumert TF. 2013. A novel monoclonal anti-CD81 antibody produced by genetic immunization efficiently inhibits hepatitis C virus cell-cell transmission. PLoS One 8:e64221. doi: 10.1371/journal.pone.0064221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA, McKelvy J, Wong-Staal F. 2011. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol 54:48–55. doi: 10.1016/j.jhep.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 165.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Calland N, Albecka A, Belouzard S, Wychowski C, Duverlie G, Descamps V, Hober D, Dubuisson J, Rouille Y, Seron K. 2012. (−)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology 55:720–729. doi: 10.1002/hep.24803. [DOI] [PubMed] [Google Scholar]
- 168.Haid S, Novodomska A, Gentzsch J, Grethe C, Geuenich S, Bankwitz D, Chhatwal P, Jannack B, Hennebelle T, Bailleul F, Keppler OT, Poenisch M, Bartenschlager R, Hernandez C, Lemasson M, Rosenberg AR, Wong-Staal F, Davioud-Charvet E, Pietschmann T. 2012. A plant-derived flavonoid inhibits entry of all HCV genotypes into human hepatocytes. Gastroenterology 143:213–222.e5. doi: 10.1053/j.gastro.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 169.Meuleman P, Albecka A, Belouzard S, Vercauteren K, Verhoye L, Wychowski C, Leroux-Roels G, Palmer KE, Dubuisson J. 2011. Griffithsin has antiviral activity against hepatitis C virus. Antimicrob Agents Chemother 55:5159–5167. doi: 10.1128/AAC.00633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Matsumura T, Hu Z, Kato T, Dreux M, Zhang YY, Imamura M, Hiraga N, Juteau JM, Cosset FL, Chayama K, Vaillant A, Liang TJ. 2009. Amphipathic DNA polymers inhibit hepatitis C virus infection by blocking viral entry. Gastroenterology 137:673–681. doi: 10.1053/j.gastro.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Marino Z, Crespo G, D'Amato M, Brambilla N, Giacovelli G, Rovati L, Costa J, Navasa M, Forns X. 2013. Intravenous silibinin monotherapy shows significant antiviral activity in HCV-infected patients in the peri-transplantation period. J Hepatol 58:415–420. doi: 10.1016/j.jhep.2012.09.034. [DOI] [PubMed] [Google Scholar]
- 172.Wagoner J, Negash A, Kane OJ, Martinez LE, Nahmias Y, Bourne N, Owen DM, Grove J, Brimacombe C, McKeating JA, Pecheur EI, Graf TN, Oberlies NH, Lohmann V, Cao F, Tavis JE, Polyak SJ. 2010. Multiple effects of silymarin on the hepatitis C virus lifecycle. Hepatology 51:1912–1921. doi: 10.1002/hep.23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liang Z, Wu S, Li Y, He L, Wu M, Jiang L, Feng L, Zhang P, Huang X. 2011. Activation of Toll-like receptor 3 impairs the dengue virus serotype 2 replication through induction of IFN-beta in cultured hepatoma cells. PLoS One 6:e23346. doi: 10.1371/journal.pone.0023346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ashkar AA, Yao XD, Gill N, Sajic D, Patrick AJ, Rosenthal KL. 2004. Toll-like receptor (TLR)-3, but not TLR4, agonist protects against genital herpes infection in the absence of inflammation seen with CpG DNA. J Infect Dis 190:1841–1849. doi: 10.1086/425079. [DOI] [PubMed] [Google Scholar]
- 175.Isogawa M, Robek MD, Furuichi Y, Chisari FV. 2005. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol 79:7269–7272. doi: 10.1128/JVI.79.11.7269-7272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Harte MT, Haga IR, Maloney G, Gray P, Reading PC, Bartlett NW, Smith GL, Bowie A, O'Neill LA. 2003. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J Exp Med 197:343–351. doi: 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sang Y, Ross CR, Rowland RR, Blecha F. 2008. Toll-like receptor 3 activation decreases porcine arterivirus infection. Viral Immunol 21:303–313. doi: 10.1089/vim.2008.0042. [DOI] [PubMed] [Google Scholar]
- 178.Sariol CA, Martinez MI, Rivera F, Rodriguez IV, Pantoja P, Abel K, Arana T, Giavedoni L, Hodara V, White LJ, Anglero YI, Montaner LJ, Kraiselburd EN. 2011. Decreased dengue replication and an increased anti-viral humoral response with the use of combined Toll-like receptor 3 and 7/8 agonists in macaques. PLoS One 6:e19323. doi: 10.1371/journal.pone.0019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Klotz D, Gerhauser I. 2019. Interferon-stimulated genes—mediators of the innate immune response during canine distemper virus infection. Int J Mol Sci 20:1620. doi: 10.3390/ijms20071620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Metz P, Reuter A, Bender S, Bartenschlager R. 2013. Interferon-stimulated genes and their role in controlling hepatitis C virus. J Hepatol 59:1331–1341. doi: 10.1016/j.jhep.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 181.Estrabaud E, Asselah T. 2010. Interferon-stimulated gene 15: a dual activity during hepatitis C virus infection. Gut 59:1017–1019. doi: 10.1136/gut.2009.206847. [DOI] [PubMed] [Google Scholar]
- 182.Kim IW, Hwang JY, Kim SK, Kim JK, Park HS. 2007. Interferon-stimulated genes response in endothelial cells following Hantaan virus infection. J Korean Med Sci 22:987–992. doi: 10.3346/jkms.2007.22.6.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Baizabal-Aguirre VM, Rosales C, López-Macías C, Gómez MI. 2016. Control and resolution mechanisms of the inflammatory response 2016. Mediators Inflamm 2016:3591797. doi: 10.1155/2016/3591797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Baizabal-Aguirre VM, Rosales C, López-Macías C, Gómez MI. 2014. Control and resolution mechanisms of the inflammatory response. Mediators Inflamm 2014:387567. doi: 10.1155/2014/387567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sato K, Ishikawa T, Okumura A, Yamauchi T, Sato S, Ayada M, Matsumoto E, Hotta N, Oohashi T, Fukuzawa Y, Kakumu S. 2007. Expression of Toll-like receptors in chronic hepatitis C virus infection. J Gastroenterol Hepatol 22:1627–1632. doi: 10.1111/j.1440-1746.2006.04783.x. [DOI] [PubMed] [Google Scholar]
- 186.Jahanban-Esfahlan R, Seidi K, Majidinia M, Karimian A, Yousefi B, Nabavi SM, Astani A, Berindan-Neagoe I, Gulei D, Fallarino F, Gargaro M, Manni G, Pirro M, Xu S, Sadeghi M, Nabavi SF, Shirooie S. 2019. Toll-like receptors as novel therapeutic targets for herpes simplex virus infection. Rev Med Virol 29:e2048. doi: 10.1002/rmv.2048. [DOI] [PubMed] [Google Scholar]
- 187.Boni C, Vecchi A, Rossi M, Laccabue D, Giuberti T, Alfieri A, Lampertico P, Grossi G, Facchetti F, Brunetto MR, Coco B, Cavallone D, Mangia A, Santoro R, Piazzolla V, Lau A, Gaggar A, Subramanian GM, Ferrari C. 2018. TLR7 agonist increases responses of hepatitis B virus-specific T cells and natural killer cells in patients with chronic hepatitis B treated with nucleos(t)ide analogues. Gastroenterology 154:1764–1777.e7. doi: 10.1053/j.gastro.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 188.Vanwalscappel B, Tada T, Landau NR. 2018. Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology 522:199–208. doi: 10.1016/j.virol.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Patel MC, Shirey KA, Pletneva LM, Boukhvalova MS, Garzino-Demo A, Vogel SN, Blanco JC. 2014. Novel drugs targeting Toll-like receptors for antiviral therapy. Future Virol 9:811–829. doi: 10.2217/fvl.14.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang F, Shen F, Wang Y, Li Z, Chen J, Yuan Z. 2020. Residues Asn118 and Glu119 of hepatitis B virus X protein are critical for HBx-mediated inhibition of RIG-I-MAVS signaling. Virology 539:92–103. doi: 10.1016/j.virol.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 191.Dalrymple NA, Cimica V, Mackow ER. 2015. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. mBio 6:e00553-15. doi: 10.1128/mBio.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. 2013. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog 9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pattabhi S, Wilkins CR, Dong R, Knoll ML, Posakony J, Kaiser S, Mire CE, Wang ML, Ireton RC, Geisbert TW, Bedard KM, Iadonato SP, Loo YM, Gale M Jr.. 2015. Targeting innate immunity for antiviral therapy through small molecule agonists of the RLR pathway. J Virol 90:2372–2387. doi: 10.1128/JVI.02202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sharma S, Sundararajan A, Suryawanshi A, Kumar N, Veiga-Parga T, Kuchroo VK, Thomas PG, Sangster MY, Rouse BT. 2011. T cell immunoglobulin and mucin protein-3 (Tim-3)/galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proc Natl Acad Sci U S A 108:19001–19006. doi: 10.1073/pnas.1107087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Velu V, Titanji K, Zhu B, Husain S, Pladevega A, Lai L, Vanderford TH, Chennareddi L, Silvestri G, Freeman GJ, Ahmed R, Amara RR. 2009. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature 458:206–210. doi: 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wykes MN, Lewin SR. 2018. Immune checkpoint blockade in infectious diseases. Nat Rev Immunol 18:91–104. doi: 10.1038/nri.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Suvas S, Kumaraguru U, Pack CD, Lee S, Rouse BT. 2003. CD4+CD25+ T cells regulate virus-specific primary and memory CD8+ T cell responses. J Exp Med 198:889–901. doi: 10.1084/jem.20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dusheiko G. 2003. Adefovir dipivoxil for the treatment of HBeAg-positive chronic hepatitis B: a review of the major clinical studies. J Hepatol 39(Suppl 1):116–123. doi: 10.1016/S0168-8278(03)00314-3. [DOI] [PubMed] [Google Scholar]
- 199.Sherman AC, Trehanpati N, Daucher M, Davey RT, Masur H, Sarin SK, Kottilil S, Kohli A. 2013. Augmentation of hepatitis B virus-specific cellular immunity with programmed death receptor-1/programmed death receptor-L1 blockade in hepatitis B virus and HIV/hepatitis B virus coinfected patients treated with adefovir. AIDS Res Hum Retroviruses 29:665–672. doi: 10.1089/aid.2012.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stoop JN, van der Molen RG, Kuipers EJ, Kusters JG, Janssen HL. 2007. Inhibition of viral replication reduces regulatory T cells and enhances the antiviral immune response in chronic hepatitis B. Virology 361:141–148. doi: 10.1016/j.virol.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 201.Trehanpati N, Vyas AK. 2017. Immune regulation by T regulatory cells in hepatitis B virus-related inflammation and cancer. Scand J Immunol 85:175–181. doi: 10.1111/sji.12524. [DOI] [PubMed] [Google Scholar]
- 202.Sehrawat S, Rouse BT. 2008. Anti-inflammatory effects of FTY720 against viral-induced immunopathology: role of drug-induced conversion of T cells to become Foxp3+ regulators. J Immunol 180:7636–7647. doi: 10.4049/jimmunol.180.11.7636. [DOI] [PubMed] [Google Scholar]
- 203.Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. 2019. Innate and adaptive immune memory: an evolutionary continuum in the host's response to pathogens. Cell Host Microbe 25:13–26. doi: 10.1016/j.chom.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 204.Mond JJ, Caporale LH, Thorbecke GJ. 1974. Kinetics of B cell memory development during a thymus “independent” immune response. Cell Immunol 10:105–116. doi: 10.1016/0008-8749(74)90155-5. [DOI] [PubMed] [Google Scholar]
- 205.Blattman JN, Grayson JM, Wherry EJ, Kaech SM, Smith KA, Ahmed R. 2003. Therapeutic use of IL-2 to enhance antiviral T-cell responses in vivo. Nat Med 9:540–547. doi: 10.1038/nm866. [DOI] [PubMed] [Google Scholar]
- 206.Phillips S, Mistry S, Riva A, Cooksley H, Hadzhiolova-Lebeau T, Plavova S, Katzarov K, Simonova M, Zeuzem S, Woffendin C, Chen PJ, Peng CY, Chang TT, Lueth S, De Knegt R, Choi MS, Wedemeyer H, Dao M, Kim CW, Chu HC, Wind-Rotolo M, Williams R, Cooney E, Chokshi S. 2017. Peg-interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front Immunol 8:621. doi: 10.3389/fimmu.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wilson EB, Brooks DG. 2011. The role of IL-10 in regulating immunity to persistent viral infections. Curr Top Microbiol Immunol 350:39–65. doi: 10.1007/82_2010_96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Teijaro JR. 2017. Cytokine storms in infectious diseases. Semin Immunopathol 39:501–503. doi: 10.1007/s00281-017-0640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Us D. 2008. Cytokine storm in avian influenza. Mikrobiyol Bul 42:365–380. (In Turkish.) [PubMed] [Google Scholar]
- 210.Gu Y, Hsu AC, Pang Z, Pan H, Zuo X, Wang G, Zheng J, Wang F. 2019. Role of the innate cytokine storm induced by the influenza A virus. Viral Immunol 32:244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- 211.Ferreira RAX, de Oliveira SA, Gandini M, Ferreira L. d C, Correa G, Abiraude FM, Reid MM, Cruz OG, Kubelka CF. 2015. Circulating cytokines and chemokines associated with plasma leakage and hepatic dysfunction in Brazilian children with dengue fever. Acta Trop 149:138–147. doi: 10.1016/j.actatropica.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 212.Patro ARK, Mohanty S, Prusty BK, Singh DK, Gaikwad S, Saswat T, Chattopadhyay S, Das BK, Tripathy R, Ravindran B. 2019. Cytokine signature associated with disease severity in dengue. Viruses 11:34. doi: 10.3390/v11010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Younan P, Iampietro M, Nishida A, Ramanathan P, Santos RI, Dutta M, Lubaki NM, Koup RA, Katze MG, Bukreyev A. 2017. Ebola virus binding to Tim-1 on T lymphocytes induces a cytokine storm. mBio 8:e00845-17. doi: 10.1128/mBio.00845-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kumar N, Sharma S, Barua S, Tripathi BN, Rouse BT. 2018. Virological and immunological outcomes of coinfections. Clin Microbiol Rev 31:e00111-17. doi: 10.1128/CMR.00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Szretter KJ, Gangappa S, Lu X, Smith C, Shieh WJ, Zaki SR, Sambhara S, Tumpey TM, Katz JM. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol 81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 217.Salomon R, Hoffmann E, Webster RG. 2007. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc Natl Acad Sci U S A 104:12479–12481. doi: 10.1073/pnas.0705289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Droebner K, Reiling SJ, Planz O. 2008. Role of hypercytokinemia in NF-kappaB p50-deficient mice after H5N1 influenza A virus infection. J Virol 82:11461–11466. doi: 10.1128/JVI.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. 2012. Into the eye of the cytokine storm. Microbiol Mol Biol Rev 76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S, Jarvik GP, Hajjar AM, Nickerson DA, Rieder M, Sevransky J, Maloney JP, Moss M, Martin G, Shanholtz C, Garcia JG, Gao L, Brower R, Barnes KC, Walley KR, Russell JA, Martin TR. 2008. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med 178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bannister S, Messina NL, Novakovic B, Curtis N. 2020. The emerging role of epigenetics in the immune response to vaccination and infection: a systematic review. Epigenetics doi: 10.1080/15592294.2020.1712814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 223.Cheedipudi S, Genolet O, Dobreva G. 2014. Epigenetic inheritance of cell fates during embryonic development. Front Genet 5:19. doi: 10.3389/fgene.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Miller TA, Witter DJ, Belvedere S. 2003. Histone deacetylase inhibitors. J Med Chem 46:5097–5116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- 225.Patil V, Guerrant W, Chen PC, Gryder B, Benicewicz DB, Khan SI, Tekwani BL, Oyelere AK. 2010. Antimalarial and antileishmanial activities of histone deacetylase inhibitors with triazole-linked cap group. Bioorg Med Chem 18:415–425. doi: 10.1016/j.bmc.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Casadesus J. 2016. Bacterial DNA methylation and methylomes. Adv Exp Med Biol 945:35–61. [DOI] [PubMed] [Google Scholar]
- 227.Bhat S, Kabekkodu SP, Noronha A, Satyamoorthy K. 2016. Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie 121:298–311. doi: 10.1016/j.biochi.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 228.Burley M, Roberts S, Parish JL. 2020. Epigenetic regulation of human papillomavirus transcription in the productive virus life cycle. Semin Immunopathol doi: 10.1007/s00281-019-00773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Knipe DM. 2015. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology 479–480:153–159. doi: 10.1016/j.virol.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhang X, Hou J, Lu M. 2013. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front Genet 4:202. doi: 10.3389/fgene.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou Q, Lin Q, Cheng D, Liao Q, Zheng L, Gong Y. 2012. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett 586:3271–3278. doi: 10.1016/j.febslet.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 232.Choi SJ, Shin YS, Kang BW, Kim JG, Won KJ, Lieberman PM, Cho H, Kang H. 2017. DNA hypermethylation induced by Epstein-Barr virus in the development of Epstein-Barr virus-associated gastric carcinoma. Arch Pharm Res 40:894–905. doi: 10.1007/s12272-017-0939-5. [DOI] [PubMed] [Google Scholar]
- 233.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lu CY, Chang YC, Hua CH, Chuang C, Huang SH, Kung SH, Hour MJ, Lin CW. 2017. Tubacin, an HDAC6 selective inhibitor, reduces the replication of the Japanese encephalitis virus via the decrease of viral RNA synthesis. Int J Mol Sci 18:954. doi: 10.3390/ijms18050954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kozlov MV, Kleymenova AA, Konduktorov KA, Malikova AZ, Kochetkov SN. 2014. Selective inhibitor of histone deacetylase 6 (tubastatin A) suppresses proliferation of hepatitis C virus replicon in culture of human hepatocytes. Biochemistry (Mosc) 79:637–642. doi: 10.1134/S0006297914070050. [DOI] [PubMed] [Google Scholar]
- 236.Sato A, Saito Y, Sugiyama K, Sakasegawa N, Muramatsu T, Fukuda S, Yoneya M, Kimura M, Ebinuma H, Hibi T, Ikeda M, Kato N, Saito H. 2013. Suppressive effect of the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) on hepatitis C virus replication. J Cell Biochem 114:1987–1996. doi: 10.1002/jcb.24541. [DOI] [PubMed] [Google Scholar]
- 237.Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, Cantor H. 2000. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science 287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 238.Danaher RJ, Jacob RJ, Steiner MR, Allen WR, Hill JM, Miller CS. 2005. Histone deacetylase inhibitors induce reactivation of herpes simplex virus type 1 in a latency-associated transcript-independent manner in neuronal cells. J Neurovirol 11:306–317. doi: 10.1080/13550280590952817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hurley EA, Thorley-Lawson DA. 1988. B cell activation and the establishment of Epstein-Barr virus latency. J Exp Med 168:2059–2075. doi: 10.1084/jem.168.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Speck SH, Chatila T, Flemington E. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol 5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 241.Ritchie D, Piekarz RL, Blombery P, Karai LJ, Pittaluga S, Jaffe ES, Raffeld M, Janik JE, Prince HM, Bates SE. 2009. Reactivation of DNA viruses in association with histone deacetylase inhibitor therapy: a case series report. Haematologica 94:1618–1622. doi: 10.3324/haematol.2009.008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhou L, He X, Gao B, Xiong S. 2015. Inhibition of histone deacetylase activity aggravates coxsackievirus B3-induced myocarditis by promoting viral replication and myocardial apoptosis. J Virol 89:10512–10523. doi: 10.1128/JVI.01028-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Li H, Tang H. 2009. On the detection of HBV cccDNA and its clinical significance: an overview of research advancement. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 26:662–666. (In Chinese.) [PubMed] [Google Scholar]
- 244.Chen Y, Sze J, He ML. 2004. HBV cccDNA in patients' sera as an indicator for HBV reactivation and an early signal of liver damage. World J Gastroenterol 10:82–85. doi: 10.3748/wjg.v10.i1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Dong J, Ying J, Qiu X, Lu Y, Zhang M. 2018. Advanced strategies for eliminating the cccDNA of HBV. Dig Dis Sci 63:7–15. doi: 10.1007/s10620-017-4842-1. [DOI] [PubMed] [Google Scholar]
- 246.Bowden S, Locarnini S, Chang TT, Chao YC, Han KH, Gish RG, de Man RA, Yu M, Llamoso C, Tang H. 2015. Covalently closed-circular hepatitis B virus DNA reduction with entecavir or lamivudine. World J Gastroenterol 21:4644–4651. doi: 10.3748/wjg.v21.i15.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wong DK, Yuen MF, Ngai VW, Fung J, Lai CL. 2006. One-year entecavir or lamivudine therapy results in reduction of hepatitis B virus intrahepatic covalently closed circular DNA levels. Antivir Ther 11:909–916. [PubMed] [Google Scholar]
- 248.Shi YM, Chu F, Shi C, He J, Chen DW, Gan Y, Wang FC, Xu ZQ, Zhong YW, Zhang XC, Zhang M. 2018. Value of quantitation of hepatitis B virus covalently closed circular DNA and HBsAg in children with chronic hepatitis B in predicting the efficacy of antiviral therapy. Zhonghua Gan Zang Bing Za Zhi 26:63–65. (In Chinese.) doi: 10.3760/cma.j.issn.1007-3418.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 249.Chen R, Lu Z, Huang L. 2014. A case report of HBsAg seroconversion with intrahepatic hepatitis B virus covalently closed circular DNA during nucleoside analog therapy. Zhonghua Gan Zang Bing Za Zhi 22:545–546. (In Chinese.) [PubMed] [Google Scholar]
- 250.Sung JJ, Wong ML, Bowden S, Liew CT, Hui AY, Wong VW, Leung NW, Locarnini S, Chan HL. 2005. Intrahepatic hepatitis B virus covalently closed circular DNA can be a predictor of sustained response to therapy. Gastroenterology 128:1890–1897. doi: 10.1053/j.gastro.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 251.Yang F. 2018. Post-translational modification control of HBV biological processes. Front Microbiol 9:2661. doi: 10.3389/fmicb.2018.02661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hong X, Kim ES, Guo H. 2017. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: implications for epigenetic therapy against chronic hepatitis B. Hepatology 66:2066–2077. doi: 10.1002/hep.29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. 2015. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci U S A 112:E5715–E5724. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lubyova B, Hodek J, Zabransky A, Prouzova H, Hubalek M, Hirsch I, Weber J. 2017. PRMT5: a novel regulator of hepatitis B virus replication and an arginine methylase of HBV core. PLoS One 12:e0186982. doi: 10.1371/journal.pone.0186982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mitra B, Thapa RJ, Guo H, Block TM. 2018. Host functions used by hepatitis B virus to complete its life cycle: implications for developing host-targeting agents to treat chronic hepatitis B. Antiviral Res 158:185–198. doi: 10.1016/j.antiviral.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lang F, Li X, Vladimirova O, Hu B, Chen G, Xiao Y, Singh V, Lu D, Li L, Han H, Wickramasinghe JM, Smith ST, Zheng C, Li Q, Lieberman PM, Fraser NW, Zhou J. 2017. CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci Rep 7:39861. doi: 10.1038/srep39861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Huang J, Kent JR, Placek B, Whelan KA, Hollow CM, Zeng PY, Fraser NW, Berger SL. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol 80:5740–5746. doi: 10.1128/JVI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Knipe DM, Lieberman PM, Jung JU, McBride AA, Morris KV, Ott M, Margolis D, Nieto A, Nevels M, Parks RJ, Kristie TM. 2013. Snapshots: chromatin control of viral infection. Virology 435:141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 260.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. 2014. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. 2013. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 4:e00558-12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tallmadge RL, Žygelytė E, Van de Walle GR, Kristie TM, Felippe MJB. 2018. Effect of a histone demethylase inhibitor on equine herpesvirus-1 activity in vitro. Front Vet Sci 5:34. doi: 10.3389/fvets.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Messer HG, Jacobs D, Dhummakupt A, Bloom DC. 2015. Inhibition of H3K27me3-specific histone demethylases JMJD3 and UTX blocks reactivation of herpes simplex virus 1 in trigeminal ganglion neurons. J Virol 89:3417–3420. doi: 10.1128/JVI.03052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Shan J, Zhao B, Shan Z, Nie J, Deng R, Xiong R, Tsun A, Pan W, Zhao H, Chen L, Jin Y, Qian Z, Lui K, Liang R, Li D, Sun B, Lavillette D, Xu K, Li B. 2017. Histone demethylase LSD1 restricts influenza A virus infection by erasing IFITM3-K88 monomethylation. PLoS Pathog 13:e1006773. doi: 10.1371/journal.ppat.1006773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sakane N, Kwon HS, Pagans S, Kaehlcke K, Mizusawa Y, Kamada M, Lassen KG, Chan J, Greene WC, Schnoelzer M, Ott M. 2011. Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog 7:e1002184. doi: 10.1371/journal.ppat.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Robertson KD. 2002. DNA methylation and chromatin—unraveling the tangled web. Oncogene 21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 268.Severin PM, Zou X, Gaub HE, Schulten K. 2011. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res 39:8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 270.Bachman KE, Rountree MR, Baylin SB. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 271.Moore LD, Le T, Fan G. 2013. DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bestor TH, Tycko B. 1996. Creation of genomic methylation patterns. Nat Genet 12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 273.Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J. 2014. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 9:e110442. doi: 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. 2003. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J Virol 77:12450–12459. doi: 10.1128/jvi.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lieberman PM. 2016. Epigenetics and genetics of viral latency. Cell Host Microbe 19:619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Toth M, Muller U, Doerfler W. 1990. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J Mol Biol 214:673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- 277.Sutter D, Doerfler W. 1980. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A 77:253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Murata T, Kondo Y, Sugimoto A, Kawashima D, Saito S, Isomura H, Kanda T, Tsurumi T. 2012. Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J Virol 86:4752–4761. doi: 10.1128/JVI.06768-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Chan AT, Tao Q, Robertson KD, Flinn IW, Mann RB, Klencke B, Kwan WH, Leung TW, Johnson PJ, Ambinder RF. 2004. Azacitidine induces demethylation of the Epstein-Barr virus genome in tumors. J Clin Oncol 22:1373–1381. doi: 10.1200/JCO.2004.04.185. [DOI] [PubMed] [Google Scholar]
- 280.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, O'Brien C, De Carvalho DD. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Paschos K, Allday MJ. 2010. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol 18:439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Adhya D, Basu A. 2010. Epigenetic modulation of host: new insights into immune evasion by viruses. J Biosci 35:647–663. doi: 10.1007/s12038-010-0072-9. [DOI] [PubMed] [Google Scholar]
- 283.Arora P, Kim EO, Jung JK, Jang KL. 2008. Hepatitis C virus core protein downregulates E-cadherin expression via activation of DNA methyltransferase 1 and 3b. Cancer Lett 261:244–252. doi: 10.1016/j.canlet.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 284.Ko E, Kim SJ, Joh JW, Park CK, Park J, Kim DH. 2008. CpG island hypermethylation of SOCS-1 gene is inversely associated with HBV infection in hepatocellular carcinoma. Cancer Lett 271:240–250. doi: 10.1016/j.canlet.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 285.Higgs MR, Lerat H, Pawlotsky JM. 2010. Downregulation of Gadd45beta expression by hepatitis C virus leads to defective cell cycle arrest. Cancer Res 70:4901–4911. doi: 10.1158/0008-5472.CAN-09-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Cicchini L, Westrich JA, Xu T, Vermeer DW, Berger JN, Clambey ET, Lee D, Song JI, Lambert PF, Greer RO, Lee JH, Pyeon D. 2016. Suppression of antitumor immune responses by human papillomavirus through epigenetic downregulation of CXCL14. mBio 7:e00270-16. doi: 10.1128/mBio.00270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Rincon-Orozco B, Halec G, Rosenberger S, Muschik D, Nindl I, Bachmann A, Ritter TM, Dondog B, Ly R, Bosch FX, Zawatzky R, Rösl F. 2009. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res 69:8718–8725. doi: 10.1158/0008-5472.CAN-09-0550. [DOI] [PubMed] [Google Scholar]
- 288.Zheng DL, Zhang L, Cheng N, Xu X, Deng Q, Teng XM, Wang KS, Zhang X, Huang J, Han ZG. 2009. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol 50:377–387. doi: 10.1016/j.jhep.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 289.Hattori N, Ushijima T. 2016. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med 8:10. doi: 10.1186/s13073-016-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhou F, Muller-Tidow C. 2019. NPM1 functions in epitranscriptomics. Nat Genet 51:1436–1437. doi: 10.1038/s41588-019-0510-z. [DOI] [PubMed] [Google Scholar]
- 291.Liu N, Pan T. 2016. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol 23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 292.Witkin KL, Hanlon SE, Strasburger JA, Coffin JM, Jaffrey SR, Howcroft TK, Dedon PC, Steitz JA, Daschner PJ, Read-Connole E. 2015. RNA editing, epitranscriptomics, and processing in cancer progression. Cancer Biol Ther 16:21–27. doi: 10.4161/15384047.2014.987555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Martin J. 2018. Epitranscriptomics: mapping methods and beyond. Biotechniques 65:121–124. doi: 10.2144/btn-2018-0117. [DOI] [PubMed] [Google Scholar]
- 294.Kennedy EM, Courtney DG, Tsai K, Cullen BR. 2017. Viral epitranscriptomics. J Virol 91:e02263-16. doi: 10.1128/JVI.02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhao L, Wientjes MG, Au JL. 2004. Evaluation of combination chemotherapy: integration of nonlinear regression, curve shift, isobologram, and combination index analyses. Clin Cancer Res 10:7994–8004. doi: 10.1158/1078-0432.CCR-04-1087. [DOI] [PubMed] [Google Scholar]
- 296.Garg V, van Heeswijk R, Lee JE, Alves K, Nadkarni P, Luo X. 2011. Effect of telaprevir on the pharmacokinetics of cyclosporine and tacrolimus. Hepatology 54:20–27. doi: 10.1002/hep.24443. [DOI] [PubMed] [Google Scholar]
- 297.Imran M, Manzoor S, Khattak NM, Khalid M, Ahmed QL, Parvaiz F, Tariq M, Ashraf J, Ashraf W, Azam S, Ashraf M. 2014. Current and future therapies for hepatitis C virus infection: from viral proteins to host targets. Arch Virol 159:831–846. doi: 10.1007/s00705-013-1803-7. [DOI] [PubMed] [Google Scholar]
- 298.Xiao F, Fofana I, Thumann C, Mailly L, Alles R, Robinet E, Meyer N, Schaeffer M, Habersetzer F, Doffoel M, Leyssen P, Neyts J, Zeisel MB, Baumert TF. 2015. Synergy of entry inhibitors with direct-acting antivirals uncovers novel combinations for prevention and treatment of hepatitis C. Gut 64:483–494. doi: 10.1136/gutjnl-2013-306155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Pizzorno A, Padey B, Terrier O, Rosa-Calatrava M. 2019. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. Front Immunol 10:531. doi: 10.3389/fimmu.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Pizzorno A, Terrier O, Nicolas de Lamballerie C, Julien T, Padey B, Traversier A, Roche M, Hamelin ME, Rheaume C, Croze S, Escuret V, Poissy J, Lina B, Legras-Lachuer C, Textoris J, Boivin G, Rosa-Calatrava M. 2019. Repurposing of drugs as novel influenza inhibitors from clinical gene expression infection signatures. Front Immunol 10:60. doi: 10.3389/fimmu.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Botta L, Rivara M, Zuliani V, Radi M. 2018. Drug repurposing approaches to fight dengue virus infection and related diseases. Front Biosci (Landmark Ed) 23:997–1019. doi: 10.2741/4630. [DOI] [PubMed] [Google Scholar]
- 302.Kang H, Kim C, Kim DE, Song JH, Choi M, Choi K, Kang M, Lee K, Kim HS, Shin JS, Kim J, Han SB, Lee MY, Lee SU, Lee CK, Kim M, Ko HJ, van Kuppeveld FJ, Cho S. 2015. Synergistic antiviral activity of gemcitabine and ribavirin against enteroviruses. Antiviral Res 124:1–10. doi: 10.1016/j.antiviral.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 303.Zhang K, Cheng L, Weir MD, Bai YX, Xu HH. 2016. Effects of quaternary ammonium chain length on the antibacterial and remineralizing effects of a calcium phosphate nanocomposite. Int J Oral Sci 8:45–53. doi: 10.1038/ijos.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng NY, Huang M, Uphadhyay AA, Gardinassi L, Petitdemange C, McCullough MP, Johnson SJ, Gill K, Cervasi B, Zou J, Bretin A, Hahn M, Gewirtz AT, Bosinger SE, Wilson PC, Li S, Alter G, Khurana S, Golding H, Pulendran B. 2019. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Dominguez-Diaz C, Garcia-Orozco A, Riera-Leal A, Padilla-Arellano JR, Fafutis-Morris M. 2019. Microbiota and its role on viral evasion: is it with us or against us? Front Cell Infect Microbiol 9:256. doi: 10.3389/fcimb.2019.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Dugger SA, Platt A, Goldstein DB. 2018. Drug development in the era of precision medicine. Nat Rev Drug Discov 17:183–196. doi: 10.1038/nrd.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Kumar N, Barua S, Thachamvally R, Tripathi BN. 2016. Systems perspective of morbillivirus replication. J Mol Microbiol Biotechnol 26:389–400. doi: 10.1159/000448842. [DOI] [PubMed] [Google Scholar]
- 308.Valenzuela-Sánchez F, Valenzuela-Méndez B, Rodríguez-Gutiérrez JF, Rello J. 2016. Personalized medicine in severe influenza. Eur J Clin Microbiol Infect Dis 35:893–897. doi: 10.1007/s10096-016-2611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Mu Y, Kodidela S, Wang Y, Kumar S, Cory TJ. 2018. The dawn of precision medicine in HIV: state of the art of pharmacotherapy. Expert Opin Pharmacother 19:1581–1595. doi: 10.1080/14656566.2018.1515916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Volberding PA. 2017. HIV treatment and prevention: an overview of recommendations from the IAS-USA Antiretroviral Guidelines Panel. Top Antivir Med 25:17–24. [PMC free article] [PubMed] [Google Scholar]
- 311.Woelk CH, Beliakova-Bethell N, Goicoechea M, Zhao Y, Du P, Rought SE, Lozach J, Perez-Santiago J, Richman DD, Smith DM, Little SJ. 2010. Gene expression before HAART initiation predicts HIV-infected individuals at risk of poor CD4+ T-cell recovery. AIDS 24:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Fellay J, Marzolini C, Meaden ER, Back DJ, Buclin T, Chave JP, Decosterd LA, Furrer H, Opravil M, Pantaleo G, Retelska D, Ruiz L, Schinkel AH, Vernazza P, Eap CB, Telenti A, Swiss HIV Cohort Study . 2002. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet 359:30–36. doi: 10.1016/S0140-6736(02)07276-8. [DOI] [PubMed] [Google Scholar]
- 313.Mukonzo JK, Roshammar D, Waako P, Andersson M, Fukasawa T, Milani L, Svensson JO, Ogwal-Okeng J, Gustafsson LL, Aklillu E. 2009. A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br J Clin Pharmacol 68:690–699. doi: 10.1111/j.1365-2125.2009.03516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbe L, Wilkinson GR, Clifford DB, D'Aquila RT, De Gruttola V, Pollard RB, Merigan TC, Hirsch MS, George AL Jr, Donahue JP, Kim RB. 2005. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult AIDS Clinical Trials Group Study. J Infect Dis 192:1931–1942. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 315.de la Tribonniere X, Broly F, Deuffic-Burban S, Bocket L, Ajana F, Viget N, Melliez H, Mouton Y, Yazdanpanah Y. 2008. ABCB1 allele polymorphism is associated with virological efficacy in naive HIV-infected patients on HAART containing nonboosted PIs but not boosted PIs. HIV Clin Trials 9:192–201. doi: 10.1310/hct0903-192. [DOI] [PubMed] [Google Scholar]
- 316.Zhu D, Taguchi-Nakamura H, Goto M, Odawara T, Nakamura T, Yamada H, Kotaki H, Sugiura W, Iwamoto A, Kitamura Y. 2004. Influence of single-nucleotide polymorphisms in the multidrug resistance-1 gene on the cellular export of nelfinavir and its clinical implication for highly active antiretroviral therapy. Antivir Ther 9:929–935. [PubMed] [Google Scholar]
- 317.Parathyras J, Gebhardt S, Hillermann-Rebello R, Grobbelaar N, Venter M, Warnich L. 2009. A pharmacogenetic study of CD4 recovery in response to HIV antiretroviral therapy in two South African population groups. J Hum Genet 54:261–265. doi: 10.1038/jhg.2009.20. [DOI] [PubMed] [Google Scholar]
- 318.Nasi M, Borghi V, Pinti M, Bellodi C, Lugli E, Maffei S, Troiano L, Richeldi L, Mussini C, Esposito R, Cossarizza A. 2003. MDR1 C3435T genetic polymorphism does not influence the response to antiretroviral therapy in drug-naive HIV-positive patients. AIDS 17:1696–1698. [DOI] [PubMed] [Google Scholar]
- 319.Brumme ZL, Dong WW, Chan KJ, Hogg RS, Montaner JS, O'Shaughnessy MV, Harrigan PR. 2003. Influence of polymorphisms within the CX3CR1 and MDR-1 genes on initial antiretroviral therapy response. AIDS 17:201–208. [DOI] [PubMed] [Google Scholar]
- 320.Hendrickson SL, Jacobson LP, Nelson GW, Phair JP, Lautenberger J, Johnson RC, Kingsley L, Margolick JB, Detels R, Goedert JJ, O'Brien SJ. 2008. Host genetic influences on highly active antiretroviral therapy efficacy and AIDS-free survival. J Acquir Immune Defic Syndr 48:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Lynn DJ, Pulendran B. 2018. The potential of the microbiota to influence vaccine responses. J Leukoc Biol 103:225–231. doi: 10.1189/jlb.5MR0617-216R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Qureshi A, Tantray VG, Kirmani AR, Ahangar AG. 2018. A review on current status of antiviral siRNA. Rev Med Virol 28:e1976. doi: 10.1002/rmv.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Hirsch AJ. 2010. The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol 5:303–311. doi: 10.2217/fmb.09.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 329.Su WC, Chen YC, Tseng CH, Hsu PW, Tung KF, Jeng KS, Lai MM. 2013. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc Natl Acad Sci U S A 110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 331.Tran AT, Rahim MN, Ranadheera C, Kroeker A, Cortens JP, Opanubi KJ, Wilkins JA, Coombs KM. 2013. Knockdown of specific host factors protects against influenza virus-induced cell death. Cell Death Dis 4:e769. doi: 10.1038/cddis.2013.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ackerman EE, Kawakami E, Katoh M, Watanabe T, Watanabe S, Tomita Y, Lopes TJ, Matsuoka Y, Kitano H, Shoemaker JE, Kawaoka Y. 2018. Network-guided discovery of influenza virus replication host factors. mBio 9:e02002-18. doi: 10.1128/mBio.02002-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Stertz S, Shaw ML. 2011. Uncovering the global host cell requirements for influenza virus replication via RNAi screening. Microbes Infect 13:516–525. doi: 10.1016/j.micinf.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, André P, Lotteau V. 2012. Genetic screens for the control of influenza virus replication: from meta-analysis to drug discovery. Mol Biosyst 8:1297–1303. doi: 10.1039/c2mb05416g. [DOI] [PubMed] [Google Scholar]
- 336.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. 2009. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Pache L, Konig R, Chanda SK. 2011. Identifying HIV-1 host cell factors by genome-scale RNAi screening. Methods 53:3–12. doi: 10.1016/j.ymeth.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 338.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 339.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 341.Kok KH, Lei T, Jin DY. 2009. siRNA and shRNA screens advance key understanding of host factors required for HIV-1 replication. Retrovirology 6:78. doi: 10.1186/1742-4690-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Zhao H, Lin W, Kumthip K, Cheng D, Fusco DN, Hofmann O, Jilg N, Tai AW, Goto K, Zhang L, Hide W, Jang JY, Peng LF, Chung RT. 2012. A functional genomic screen reveals novel host genes that mediate interferon-alpha's effects against hepatitis C virus. J Hepatol 56:326–333. doi: 10.1016/j.jhep.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Cao D, Haussecker D, Huang Y, Kay MA. 2009. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA 15:1971–1979. doi: 10.1261/rna.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Lavanya M, Cuevas CD, Thomas M, Cherry S, Ross SR. 2013. siRNA screen for genes that affect Junin virus entry uncovers voltage-gated calcium channels as a therapeutic target. Sci Transl Med 5:204ra131. doi: 10.1126/scitranslmed.3006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Lee AS, Burdeinick-Kerr R, Whelan SP. 2014. A genome-wide small interfering RNA screen identifies host factors required for vesicular stomatitis virus infection. J Virol 88:8355–8360. doi: 10.1128/JVI.00642-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Martin S, Chiramel AI, Schmidt ML, Chen YC, Whitt N, Watt A, Dunham EC, Shifflett K, Traeger S, Leske A, Buehler E, Martellaro C, Brandt J, Wendt L, Muller A, Peitsch S, Best SM, Stech J, Finke S, Romer-Oberdorfer A, Groseth A, Feldmann H, Hoenen T. 2018. A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med 10:58. doi: 10.1186/s13073-018-0570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F, Gonzalez O, Friedel CC, Barry G, Martin K, Craigon MH, Chen R, Kaza LN, Fossum E, Fazakerley JK, Efstathiou S, Volpi A, Zimmer R, Ghazal P, Haas J. 2013. A systematic analysis of host factors reveals a Med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog 9:e1003514. doi: 10.1371/journal.ppat.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Sieczkarski SB, Whittaker GR. 2003. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333–343. doi: 10.1034/j.1600-0854.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 352.Sun E, He J, Zhuang X. 2013. Dissecting the role of COPI complexes in influenza virus infection. J Virol 87:2673–2685. doi: 10.1128/JVI.02277-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Guinea R, Carrasco L. 1995. Requirement for vacuolar proton-ATPase activity during entry of influenza virus into cells. J Virol 69:2306–2312. doi: 10.1128/JVI.69.4.2306-2312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Chen J, Huang S, Chen Z. 2010. Human cellular protein nucleoporin hNup98 interacts with influenza A virus NS2/nuclear export protein and overexpression of its GLFG repeat domain can inhibit virus propagation. J Gen Virol 91:2474–2484. doi: 10.1099/vir.0.022681-0. [DOI] [PubMed] [Google Scholar]
- 355.Schneider J, Wolff T. 2009. Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine 27:6312–6316. doi: 10.1016/j.vaccine.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 356.Chou YC, Lai MM, Wu YC, Hsu NC, Jeng KS, Su WC. 2015. Variations in genome-wide RNAi screens: lessons from influenza research. J Clin Bioinforma 5:2. doi: 10.1186/s13336-015-0017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Perreira JM, Meraner P, Brass AL. 2016. Functional genomic strategies for elucidating human-virus interactions: will CRISPR knockout RNAi and haploid cells? Adv Virus Res 94:1–51. doi: 10.1016/bs.aivir.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Heaton BE, Kennedy EM, Dumm RE, Harding AT, Sacco MT, Sachs D, Heaton NS. 2017. A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep 20:1503–1512. doi: 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, Lee Y, Andrade J, tenOever B, Manicassamy B. 2018. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep 23:596–607. doi: 10.1016/j.celrep.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, Guo Z, Green S, Kowalik TF, Brass AL. 2016. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep 16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 361.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, Xing C, Schoggins JW. 2018. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol 3:1214–1223. doi: 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, Diamond MS. 2016. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi JG, Shi G, Qi L, Manjunath N, Wu H. 2015. A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep 12:673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.OhAinle M, Helms L, Vermeire J, Roesch F, Humes D, Basom R, Delrow JJ, Overbaugh J, Emerman M. 2018. A virus-packageable CRISPR screen identifies host factors mediating interferon inhibition of HIV. Elife 7:e39823. doi: 10.7554/eLife.39823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, Schumann K, Yu H, Krupzcak KM, Garcia-Beltran W, Piechocka-Trocha A, Krogan NJ, Marson A, Sabatini DM, Lander ES, Hacohen N, Walker BD. 2017. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet 49:193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Orchard RC, Wilen CB, Doench JG, Baldridge MT, McCune BT, Lee YC, Lee S, Pruett-Miller SM, Nelson CA, Fremont DH, Virgin HW. 2016. Discovery of a proteinaceous cellular receptor for a norovirus. Science 353:933–936. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A 108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Krumm SA, Ndungu JM, Yoon JJ, Dochow M, Sun A, Natchus M, Snyder JP, Plemper RK. 2011. Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS One 6:e20069. doi: 10.1371/journal.pone.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Hopcraft SE, Evans MJ. 2015. Selection of a hepatitis C virus with altered entry factor requirements reveals a genetic interaction between the E1 glycoprotein and claudins. Hepatology 62:1059–1069. doi: 10.1002/hep.27815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Dhungel P, Cao S, Yang Z. 2017. The 5'-poly(A) leader of poxvirus mRNA confers a translational advantage that can be achieved in cells with impaired cap-dependent translation. PLoS Pathog 13:e1006602. doi: 10.1371/journal.ppat.1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Puyang X, Poulin DL, Mathy JE, Anderson LJ, Ma S, Fang Z, Zhu S, Lin K, Fujimoto R, Compton T, Wiedmann B. 2010. Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors. Antimicrob Agents Chemother 54:1981–1987. doi: 10.1128/AAC.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Xiao X, Lei X, Zhang Z, Ma Y, Qi J, Wu C, Xiao Y, Li L, He B, Wang J. 2017. Enterovirus 3A facilitates viral replication by promoting phosphatidylinositol 4-kinase IIIbeta-ACBD3 interaction. J Virol 91:e00791-17. doi: 10.1128/JVI.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Arita M. 2016. Mechanism of poliovirus resistance to host phosphatidylinositol-4 kinase III beta inhibitor. ACS Infect Dis 2:140–148. doi: 10.1021/acsinfecdis.5b00122. [DOI] [PubMed] [Google Scholar]
- 375.van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Purstinger G, de Vries E, de Haan CA, Neyts J, van Kuppeveld FJ. 2012. Coxsackievirus mutants that can bypass host factor PI4KIIIbeta and the need for high levels of PI4P lipids for replication. Cell Res 22:1576–1592. doi: 10.1038/cr.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Chatterji U, Bobardt MD, Lim P, Gallay PA. 2010. Cyclophilin A-independent recruitment of NS5A and NS5B into hepatitis C virus replication complexes. J Gen Virol 91:1189–1193. doi: 10.1099/vir.0.018531-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Coelmont L, Kaptein S, Paeshuyse J, Vliegen I, Dumont JM, Vuagniaux G, Neyts J. 2009. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob Agents Chemother 53:967–976. doi: 10.1128/AAC.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Goldhill DH, Turner PE. 2014. The evolution of life history trade-offs in viruses. Curr Opin Virol 8:79–84. doi: 10.1016/j.coviro.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 379.Chantranupong L, Heineman RH. 2012. A common, non-optimal phenotypic endpoint in experimental adaptations of bacteriophage lysis time. BMC Evol Biol 12:37. doi: 10.1186/1471-2148-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Heineman RH, Bull JJ. 2007. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution 61:1695–1709. doi: 10.1111/j.1558-5646.2007.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Wang IN. 2006. Lysis timing and bacteriophage fitness. Genetics 172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Abedon ST, Hyman P, Thomas C. 2003. Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl Environ Microbiol 69:7499–7506. doi: 10.1128/AEM.69.12.7499-7506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Abedon ST. 1989. Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microb Ecol 18:79–88. doi: 10.1007/BF02030117. [DOI] [PubMed] [Google Scholar]
- 384.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 385.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 386.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 387.Wahl LM, Nowak MA. 2000. Adherence and drug resistance: predictions for therapy outcome. Proc Biol Sci 267:835–843. doi: 10.1098/rspb.2000.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Neagu IA, Olejarz J, Freeman M, Rosenbloom DIS, Nowak MA, Hill AL. 2018. Life cycle synchronization is a viral drug resistance mechanism. PLoS Comput Biol 14:e1005947. doi: 10.1371/journal.pcbi.1005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Lin K, Gallay P. 2013. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antiviral Res 99:68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Rustgi VK, Lee WM, Lawitz E, Gordon SC, Afdhal N, Poordad F, Bonkovsky HL, Bengtsson L, Chandorkar G, Harding M, McNair L, Aalyson M, Alam J, Kauffman R, Gharakhanian S, McHutchison JG, MErimepodib TRiple cOmbination Study Group . 2009. Merimepodib, pegylated interferon, and ribavirin in genotype 1 chronic hepatitis C pegylated interferon and ribavirin nonresponders. Hepatology 50:1719–1726. doi: 10.1002/hep.23204. [DOI] [PubMed] [Google Scholar]
- 391.Rustgi V, Nelson DR, Balan V, Abelson RD, Fiscella M, Migone TS, Pulkstenis E, Subramanian GM. 2009. Changes in B-lymphocyte stimulator protein levels during treatment with albinterferon alfa-2b in patients with chronic hepatitis C who have failed previous interferon therapy. Hepatol Res 39:455–462. doi: 10.1111/j.1872-034X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 392.McHutchison JG, Bartenschlager R, Patel K, Pawlotsky JM. 2006. The face of future hepatitis C antiviral drug development: recent biological and virologic advances and their translation to drug development and clinical practice. J Hepatol 44:411–421. doi: 10.1016/j.jhep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 393.Ikeda M, Abe K, Yamada M, Dansako H, Naka K, Kato N. 2006. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology 44:117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 394.Rustgi VK. 2009. Albinterferon alfa-2b, a novel fusion protein of human albumin and human interferon alfa-2b, for chronic hepatitis C. Curr Med Res Opin 25:991–1002. doi: 10.1185/03007990902779186. [DOI] [PubMed] [Google Scholar]
- 395.O'Leary JG, Chan JL, McMahon CM, Chung RT. 2007. Atorvastatin does not exhibit antiviral activity against HCV at conventional doses: a pilot clinical trial. Hepatology 45:895–898. doi: 10.1002/hep.21554. [DOI] [PubMed] [Google Scholar]
- 396.Bader T, Fazili J, Madhoun M, Aston C, Hughes D, Rizvi S, Seres K, Hasan M. 2008. Fluvastatin inhibits hepatitis C replication in humans. Am J Gastroenterol 103:1383–1389. doi: 10.1111/j.1572-0241.2008.01876.x. [DOI] [PubMed] [Google Scholar]
- 397.Sezaki H, Suzuki F, Akuta N, Yatsuji H, Hosaka T, Kobayashi M, Suzuki Y, Arase Y, Ikeda K, Miyakawa Y, Kumada H. 2009. An open pilot study exploring the efficacy of fluvastatin, pegylated interferon and ribavirin in patients with hepatitis C virus genotype 1b in high viral loads. Intervirology 52:43–48. doi: 10.1159/000213504. [DOI] [PubMed] [Google Scholar]
- 398.Forde KA, Law C, O’Flynn R, Kaplan DE. 2009. Do statins reduce hepatitis C RNA titers during routine clinical use? World J Gastroenterol 15:5020–5027. doi: 10.3748/wjg.15.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Milazzo L, Meroni L, Galazzi M, Cesari M, Caramma I, Marchetti G, Galli M, Antinori S. 2009. Does fluvastatin favour HCV replication in vivo? A pilot study on HIV-HCV coinfected patients. J Viral Hepat 16:479–484. doi: 10.1111/j.1365-2893.2009.01104.x. [DOI] [PubMed] [Google Scholar]
- 400.Lamarche MJ, Borawski J, Bose A, Capacci-Daniel C, Colvin R, Dennehy M, Ding J, Dobler M, Drumm J, Gaither LA, Gao J, Jiang X, Lin K, McKeever U, Puyang X, Raman P, Thohan S, Tommasi R, Wagner K, Xiong X, Zabawa T, Zhu S, Wiedmann B. 2012. Anti-hepatitis C virus activity and toxicity of type III phosphatidylinositol-4-kinase beta inhibitors. Antimicrob Agents Chemother 56:5149–5156. doi: 10.1128/AAC.00946-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.de Chassey B, Meyniel-Schicklin L, Vonderscher J, André P, Lotteau V. 2014. Virus-host interactomics: new insights and opportunities for antiviral drug discovery. Genome Med 6:115. doi: 10.1186/s13073-014-0115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Zeisel MB, Lupberger J, Fofana I, Baumert TF. 2013. Host-targeting agents for prevention and treatment of chronic hepatitis C—perspectives and challenges. J Hepatol 58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 403.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabárbara P, Seymour L, National Cancer Institute of Canada Clinical Trials Group . 2005. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 404.Henrich TJ, Kuritzkes DR. 2013. HIV-1 entry inhibitors: recent development and clinical use. Curr Opin Virol 3:51–57. doi: 10.1016/j.coviro.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Moss RB, Hansen C, Sanders RL, Hawley S, Li T, Steigbigel RT. 2012. A phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J Infect Dis 206:1844–1851. doi: 10.1093/infdis/jis622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Salvatore M, Satlin MJ, Jacobs SE, Jenkins SG, Schuetz AN, Moss RB, Van Besien K, Shore T, Soave R. 2016. DAS181 for treatment of parainfluenza virus infections in hematopoietic stem cell transplant recipients at a single center. Biol Blood Marrow Transplant 22:965–970. doi: 10.1016/j.bbmt.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 407.Waghmare A, Wagner T, Andrews R, Smith S, Kuypers J, Boeckh M, Moss R, Englund JA. 2015. Successful treatment of parainfluenza virus respiratory tract infection with DAS181 in 4 immunocompromised children. J Pediatric Infect Dis Soc 4:114–118. doi: 10.1093/jpids/piu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Jones BG, Hayden RT, Hurwitz JL. 2013. Inhibition of primary clinical isolates of human parainfluenza virus by DAS181 in cell culture and in a cotton rat model. Antiviral Res 100:562–566. doi: 10.1016/j.antiviral.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Triana-Baltzer GB, Babizki M, Chan MC, Wong AC, Aschenbrenner LM, Campbell ER, Li QX, Chan RW, Peiris JS, Nicholls JM, Fang F. 2010. DAS181, a sialidase fusion protein, protects human airway epithelium against influenza virus infection: an in vitro pharmacodynamic analysis. J Antimicrob Chemother 65:275–284. doi: 10.1093/jac/dkp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Chan RW, Chan MC, Wong AC, Karamanska R, Dell A, Haslam SM, Sihoe AD, Chui WH, Triana-Baltzer G, Li Q, Peiris JS, Fang F, Nicholls JM. 2009. DAS181 inhibits H5N1 influenza virus infection of human lung tissues. Antimicrob Agents Chemother 53:3935–3941. doi: 10.1128/AAC.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Zhang H. 2009. DAS181 and H5N1 virus infection. J Infect Dis 199:1250. doi: 10.1086/597479. (Letter.) (Reply, 199:1250–1251, doi:10.1086/597480.) [DOI] [PubMed] [Google Scholar]
- 412.Liu S, DeLalio LJ, Isakson BE, Wang TT. 2016. AXL-mediated productive infection of human endothelial cells by Zika virus. Circ Res 119:1183–1189. doi: 10.1161/CIRCRESAHA.116.309866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Persaud M, Martinez-Lopez A, Buffone C, Porcelli SA, Diaz-Griffero F. 2018. Infection by Zika viruses requires the transmembrane protein AXL, endocytosis and low pH. Virology 518:301–312. doi: 10.1016/j.virol.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Varghese FS, Rausalu K, Hakanen M, Saul S, Kummerer BM, Susi P, Merits A, Ahola T. 2017. Obatoclax inhibits alphavirus membrane fusion by neutralizing the acidic environment of endocytic compartments. Antimicrob Agents Chemother 61:e02227-16. doi: 10.1128/AAC.02227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Haviernik J, Stefanik M, Fojtikova M, Kali S, Tordo N, Rudolf I, Hubalek Z, Eyer L, Ruzek D. 2018. Arbidol (umifenovir): a broad-spectrum antiviral drug that inhibits medically important arthropod-borne flaviviruses. Viruses 10:E184. doi: 10.3390/v10040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Jurgeit A, McDowell R, Moese S, Meldrum E, Schwendener R, Greber UF. 2012. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog 8:e1002976. doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Munoz G, McGrath EL, Urrabaz-Garza R, Gao J, Wu P, Menon R, Saade G, Fernandez-Salas I, Rossi SL, Vasilakis N, Routh A, Bradrick SS, Garcia-Blanco MA. 2016. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Wolkerstorfer A, Kurz H, Bachhofner N, Szolar OH. 2009. Glycyrrhizin inhibits influenza A virus uptake into the cell. Antiviral Res 83:171–178. doi: 10.1016/j.antiviral.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Müller KH, Kainov DE, El Bakkouri K, Saelens X, De Brabander JK, Kittel C, Samm E, Muller CP. 2011. The proton translocation domain of cellular vacuolar ATPase provides a target for the treatment of influenza A virus infections. Br J Pharmacol 164:344–357. doi: 10.1111/j.1476-5381.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Bimbo LM, Denisova OV, Mäkilä E, Kaasalainen M, De Brabander JK, Hirvonen J, Salonen J, Kakkola L, Kainov D, Santos HA. 2013. Inhibition of influenza A virus infection in vitro by saliphenylhalamide-loaded porous silicon nanoparticles. ACS Nano 7:6884–6893. doi: 10.1021/nn402062f. [DOI] [PubMed] [Google Scholar]
- 421.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, Porotto M, Honko AN, Damoiseaux R, Miller JP, Woodson SE, Chantasirivisal S, Fontanes V, Negrete OA, Krogstad P, Dasgupta A, Moscona A, Hensley LE, Whelan SP, Faull KF, Holbrook MR, Jung ME, Lee B. 2010. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A 107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Mues MB, Cheshenko N, Wilson DW, Gunther-Cummins L, Herold BC. 2015. Dynasore disrupts trafficking of herpes simplex virus proteins. J Virol 89:6673–6684. doi: 10.1128/JVI.00636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Anantpadma M, Kouznetsova J, Wang H, Huang R, Kolokoltsov A, Guha R, Lindstrom AR, Shtanko O, Simeonov A, Maloney DJ, Maury W, LaCount DJ, Jadhav A, Davey RA. 2016. Large-scale screening and identification of novel Ebola virus and Marburg virus entry inhibitors. Antimicrob Agents Chemother 60:4471–4481. doi: 10.1128/AAC.00543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Root CN, Wills EG, McNair LL, Whittaker GR. 2000. Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J Gen Virol 81:2697–2705. doi: 10.1099/0022-1317-81-11-2697. [DOI] [PubMed] [Google Scholar]
- 425.Blazquez AB, Vazquez-Calvo A, Martin-Acebes MA, Saiz JC. 2018. Pharmacological inhibition of protein kinase C reduces West Nile virus replication. Viruses 10:91. doi: 10.3390/v10020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Harada S, Yokomizo K, Monde K, Maeda Y, Yusa K. 2007. A broad antiviral neutral glycolipid, fattiviracin FV‐8, is a membrane fluidity modulator. Cell Microbiol 9:196–203. doi: 10.1111/j.1462-5822.2006.00781.x. [DOI] [PubMed] [Google Scholar]
- 427.Ehrhardt C, Rückle A, Hrincius ER, Haasbach E, Anhlan D, Ahmann K, Banning C, Reiling SJ, Kühn J, Strobl S. 2013. The NF‐κ B inhibitor SC 75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell Microbiol 15:1198–1211. doi: 10.1111/cmi.12108. [DOI] [PubMed] [Google Scholar]
- 428.Lee M, Kim K, Park K, Kim J. 1996. Evaluation of anti-influenza effects of camostat in mice infected with non-adapted human influenza viruses. Arch Virol 141:1979–1989. doi: 10.1007/bf01718208. [DOI] [PubMed] [Google Scholar]
- 429.Kalin S, Amstutz B, Gastaldelli M, Wolfrum N, Boucke K, Havenga M, DiGennaro F, Liska N, Hemmi S, Greber UF. 2010. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J Virol 84:5336–5350. doi: 10.1128/JVI.02494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Harrison DN, Gazina EV, Purcell DF, Anderson DA, Petrou S. 2008. Amiloride derivatives inhibit coxsackievirus B3 RNA replication. J Virol 82:1465–1473. doi: 10.1128/JVI.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Yang S, Xu M, Lee EM, Gorshkov K, Shiryaev SA, He S, Sun W, Cheng YS, Hu X, Tharappel AM, Lu B, Pinto A, Farhy C, Huang CT, Zhang Z, Zhu W, Wu Y, Zhou Y, Song G, Zhu H, Shamim K, Martinez-Romero C, Garcia-Sastre A, Preston RA, Jayaweera DT, Huang R, Huang W, Xia M, Simeonov A, Ming G, Qiu X, Terskikh AV, Tang H, Song H, Zheng W. 2018. Emetine inhibits Zika and Ebola virus infections through two molecular mechanisms: inhibiting viral replication and decreasing viral entry. Cell Discov 4:31. doi: 10.1038/s41421-018-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Khandelwal N, Chander Y, Rawat KD, Riyesh T, Nishanth C, Sharma S, Jindal N, Tripathi BN, Barua S, Kumar N. 2017. Emetine inhibits replication of RNA and DNA viruses without generating drug-resistant virus variants. Antiviral Res 144:196–204. doi: 10.1016/j.antiviral.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 433.Rausch K, Hackett BA, Weinbren NL, Reeder SM, Sadovsky Y, Hunter CA, Schultz DC, Coyne CB, Cherry S. 2017. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep 18:804–815. doi: 10.1016/j.celrep.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Matsui K, Ozawa M, Kiso M, Yamashita M, Maekawa T, Kubota M, Sugano S, Kawaoka Y. 2018. Stimulation of alpha2-adrenergic receptors impairs influenza virus infection. Sci Rep 8:4631. doi: 10.1038/s41598-018-22927-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Qing J, Wang Y, Sun Y, Huang J, Yan W, Wang J, Su D, Ni C, Li J, Rao Z, Liu L, Lou Z. 2014. Cyclophilin A associates with enterovirus-71 virus capsid and plays an essential role in viral infection as an uncoating regulator. PLoS Pathog 10:e1004422. doi: 10.1371/journal.ppat.1004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Reeves PM, Bommarius B, Lebeis S, McNulty S, Christensen J, Swimm A, Chahroudi A, Chavan R, Feinberg MB, Veach D, Bornmann W, Sherman M, Kalman D. 2005. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat Med 11:731–739. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- 437.Swimm AI, Bornmann W, Jiang M, Imperiale MJ, Lukacher AE, Kalman D. 2010. Abl family tyrosine kinases regulate sialylated ganglioside receptors for polyomavirus. J Virol 84:4243–4251. doi: 10.1128/JVI.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Ullah H, Hou W, Dakshanamurthy S, Tang Q. 2019. Host targeted antiviral (HTA): functional inhibitor compounds of scaffold protein RACK1 inhibit herpes simplex virus proliferation. Oncotarget 10:3209–3226. doi: 10.18632/oncotarget.26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Stöhr S, Costa R, Sandmann L, Westhaus S, Pfaender S, Anggakusuma, Dazert E, Meuleman P, Vondran FWR, Manns MP, Steinmann E, von Hahn T, Ciesek S. 2016. Host cell mTORC1 is required for HCV RNA replication. Gut 65:2017–2028. doi: 10.1136/gutjnl-2014-308971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Lenarcic EM, Ziehr B, De Leon G, Mitchell D, Moorman NJ. 2014. Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. J Virol 88:1473–1483. doi: 10.1128/JVI.02321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Ji WT, Wang YC, Lin FL, Liao MH, Shih WL, Liu HJ. 2011. Inhibitors of phosphatidylinositol 3-kinase and mTOR but not Akt enhance replication of bovine ephemeral fever virus. Vet J 187:119–123. doi: 10.1016/j.tvjl.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 442.Elgner F, Sabino C, Basic M, Ploen D, Grunweller A, Hildt E. 2018. Inhibition of Zika virus replication by silvestrol. Viruses 10:E149. doi: 10.3390/v10040149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18:660–672. doi: 10.1101/gad.1185304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79:8057–8064. doi: 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, Pelletier J. 2011. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85:6381–6389. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Qian S, Fan W, Qian P, Zhang D, Wei Y, Chen H, Li X. 2015. Apigenin restricts FMDV infection and inhibits viral IRES driven translational activity. Viruses 7:1613–1626. doi: 10.3390/v7041613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Zhang W, Qiao H, Lv Y, Wang J, Chen X, Hou Y, Tan R, Li E. 2014. Apigenin inhibits enterovirus-71 infection by disrupting viral RNA association with trans-acting factors. PLoS One 9:e110429. doi: 10.1371/journal.pone.0110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Stantchev TS, Markovic I, Telford WG, Clouse KA, Broder CC. 2007. The tyrosine kinase inhibitor genistein blocks HIV-1 infection in primary human macrophages. Virus Res 123:178–189. doi: 10.1016/j.virusres.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Herget T, Freitag M, Morbitzer M, Kupfer R, Stamminger T, Marschall M. 2004. Novel chemical class of pUL97 protein kinase-specific inhibitors with strong anticytomegaloviral activity. Antimicrob Agents Chemother 48:4154–4162. doi: 10.1128/AAC.48.11.4154-4162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Langhammer S, Koban R, Yue C, Ellerbrok H. 2011. Inhibition of poxvirus spreading by the anti-tumor drug Gefitinib (Iressa). Antiviral Res 89:64–70. doi: 10.1016/j.antiviral.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 452.Götz V, Magar L, Dornfeld D, Giese S, Pohlmann A, Höper D, Kong B-W, Jans DA, Beer M, Haller O. 2016. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci Rep 6:23138. doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Perwitasari O, Johnson S, Yan X, Howerth E, Shacham S, Landesman Y, Baloglu E, McCauley D, Tamir S, Tompkins SM. 2014. Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza A virus replication in vitro and in vivo. J Virol 88:10228–10243. doi: 10.1128/JVI.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Watanabe K, Takizawa N, Katoh M, Hoshida K, Kobayashi N, Nagata K. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res 77:31–42. doi: 10.1016/s0168-1702(01)00263-5. [DOI] [PubMed] [Google Scholar]
- 455.Chutiwitoonchai N, Mano T, Kakisaka M, Sato H, Kondoh Y, Osada H, Kotani O, Yokoyama M, Sato H, Aida Y. 2017. Inhibition of CRM1-mediated nuclear export of influenza A nucleoprotein and nuclear export protein as a novel target for antiviral drug development. Virology 507:32–39. doi: 10.1016/j.virol.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Holzberg M, Boergeling Y, Schrader T, Ludwig S, Ehrhardt C. 2017. Vemurafenib limits influenza A virus propagation by targeting multiple signaling pathways. Front Microbiol 8:2426. doi: 10.3389/fmicb.2017.02426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Cai Y, Liu Y, Zhang X. 2007. Suppression of coronavirus replication by inhibition of the MEK signaling pathway. J Virol 81:446–456. doi: 10.1128/JVI.01705-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Colao I, Pennisi R, Venuti A, Nygardas M, Heikkila O, Hukkanen V, Sciortino MT. 2017. The ERK-1 function is required for HSV-1-mediated G1/S progression in HEP-2 cells and contributes to virus growth. Sci Rep 7:9176. doi: 10.1038/s41598-017-09529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Kim Y, Lee C. 2015. Extracellular signal-regulated kinase (ERK) activation is required for porcine epidemic diarrhea virus replication. Virology 484:181–193. doi: 10.1016/j.virol.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Ludwig S, Wolff T, Ehrhardt C, Wurzer WJ, Reinhardt J, Planz O, Pleschka S. 2004. MEK inhibition impairs influenza B virus propagation without emergence of resistant variants. FEBS Lett 561:37–43. doi: 10.1016/S0014-5793(04)00108-5. [DOI] [PubMed] [Google Scholar]
- 461.Moser LA, Schultz-Cherry S. 2008. Suppression of astrovirus replication by an ERK1/2 inhibitor. J Virol 82:7475–7482. doi: 10.1128/JVI.02193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Planz O, Pleschka S, Ludwig S. 2001. MEK-specific inhibitor U0126 blocks spread of Borna disease virus in cultured cells. J Virol 75:4871–4877. doi: 10.1128/JVI.75.10.4871-4877.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Rodriguez ME, Brunetti JE, Wachsman MB, Scolaro LA, Castilla V. 2014. Raf/MEK/ERK pathway activation is required for Junin virus replication. J Gen Virol 95:799–805. doi: 10.1099/vir.0.061242-0. [DOI] [PubMed] [Google Scholar]
- 464.Haasbach E, Muller C, Ehrhardt C, Schreiber A, Pleschka S, Ludwig S, Planz O. 2017. The MEK-inhibitor CI-1040 displays a broad anti-influenza virus activity in vitro and provides a prolonged treatment window compared to standard of care in vivo. Antiviral Res 142:178–184. doi: 10.1016/j.antiviral.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 465.Sreekanth GP, Chuncharunee A, Sirimontaporn A, Panaampon J, Srisawat C, Morchang A, Malakar S, Thuwajit P, Kooptiwut S, Suttitheptumrong A, Songprakhon P, Noisakran S, Yenchitsomanus PT, Limjindaporn T. 2014. Role of ERK1/2 signaling in dengue virus-induced liver injury. Virus Res 188:15–26. doi: 10.1016/j.virusres.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 466.Voss K, Amaya M, Mueller C, Roberts B, Kehn-Hall K, Bailey C, Petricoin E III, Narayanan A. 2014. Inhibition of host extracellular signal-regulated kinase (ERK) activation decreases New World alphavirus multiplication in infected cells. Virology 468-470:490–503. doi: 10.1016/j.virol.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Hirasawa K, Kim A, Han HS, Han J, Jun HS, Yoon JW. 2003. Effect of p38 mitogen-activated protein kinase on the replication of encephalomyocarditis virus. J Virol 77:5649–5656. doi: 10.1128/jvi.77.10.5649-5656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Zhang H, Niu X, Qian Z, Qian J, Xuan B. 2015. The c-Jun N-terminal kinase inhibitor SP600125 inhibits human cytomegalovirus replication. J Med Virol 87:2135–2144. doi: 10.1002/jmv.24286. [DOI] [PubMed] [Google Scholar]
- 469.Nacken W, Ehrhardt C, Ludwig S. 2012. Small molecule inhibitors of the c-Jun N-terminal kinase (JNK) possess antiviral activity against highly pathogenic avian and human pandemic influenza A viruses. Biol Chem 393:525–534. doi: 10.1515/hsz-2011-0270. [DOI] [PubMed] [Google Scholar]
- 470.Hu J, Ma L, Wang H, Yan H, Zhang D, Li Z, Jiang J, Li Y. 2017. A novel benzo-heterocyclic amine derivative N30 inhibits influenza virus replication by depression of inosine-5'-monophospate dehydrogenase activity. Virol J 14:55. doi: 10.1186/s12985-017-0724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Markland W, McQuaid TJ, Jain J, Kwong AD. 2000. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob Agents Chemother 44:859–866. doi: 10.1128/aac.44.4.859-866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Paeshuyse J, Dallmeier K, Neyts J. 2011. Ribavirin for the treatment of chronic hepatitis C virus infection: a review of the proposed mechanisms of action. Curr Opin Virol 1:590–598. doi: 10.1016/j.coviro.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 473.Huang JT, Tseng CP, Liao MH, Lu SC, Yeh WZ, Sakamoto N, Chen CM, Cheng JC. 2013. Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase. J Virol 87:4994–5004. doi: 10.1128/JVI.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Zu M, Li C, Fang J-S, Lian W-W, Liu A-L, Zheng L-S, Du G-H. 2015. Drug discovery of host CLK1 inhibitors for influenza treatment. Molecules 20:19735–19747. doi: 10.3390/molecules201119653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Yángüez E, Castello A, Welnowska E, Carrasco L, Goodfellow I, Nieto A. 2011. Functional impairment of eIF4A and eIF4G factors correlates with inhibition of influenza virus mRNA translation. Virology 413:93–102. doi: 10.1016/j.virol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 476.Malina A, Mills JR, Pelletier J. 2012. Emerging therapeutics targeting mRNA translation. Cold Spring Harb Perspect Biol 4:a012377. doi: 10.1101/cshperspect.a012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Huang HI, Chio CC, Lin JY. 2018. Inhibition of EV71 by curcumin in intestinal epithelial cells. PLoS One 13:e0191617. doi: 10.1371/journal.pone.0191617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Dutta K, Ghosh D, Basu A. 2009. Curcumin protects neuronal cells from Japanese encephalitis virus-mediated cell death and also inhibits infective viral particle formation by dysregulation of ubiquitin-proteasome system. J Neuroimmune Pharmacol 4:328–337. doi: 10.1007/s11481-009-9158-2. [DOI] [PubMed] [Google Scholar]
- 479.Narayanan A, Kehn-Hall K, Senina S, Lundberg L, Van Duyne R, Guendel I, Das R, Baer A, Bethel L, Turell M, Hartman AL, Das B, Bailey C, Kashanchi F. 2012. Curcumin inhibits Rift Valley fever virus replication in human cells. J Biol Chem 287:33198–33214. doi: 10.1074/jbc.M112.356535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Kim K, Kim KH, Kim HY, Cho HK, Sakamoto N, Cheong J. 2010. Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS Lett 584:707–712. doi: 10.1016/j.febslet.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 481.Bauer L, Lyoo H, van der Schaar HM, Strating JR, van Kuppeveld FJ. 2017. Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr Opin Virol 24:1–8. doi: 10.1016/j.coviro.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.van der Linden L, Wolthers KC, van Kuppeveld FJ. 2015. Replication and inhibitors of enteroviruses and parechoviruses. Viruses 7:4529–4562. doi: 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Erken R, Stelma F, Roy E, Diane S, Andre P, Vonderscher J, Eric M, Tim S, Philippe P, Christian L, Scalfaro P, Reesink H, Sousa CM, Jacob E. 2018. First clinical evaluation in chronic hepatitis B patients of the synthetic farnesoid X receptor agonist EYP001. J Hepatol 68:S488–S489. doi: 10.1016/S0168-8278(18)31226-1. [DOI] [Google Scholar]
- 484.Pan W, Luo Q, Yan X, Yuan L, Yi H, Zhang L, Li B, Zhang Y, Sun J, Qiu MZ, Yang DJ. 2018. A novel SMAC mimetic APG-1387 exhibits dual antitumor effect on HBV-positive hepatocellular carcinoma with high expression of cIAP2 by inducing apoptosis and enhancing innate anti-tumor immunity. Biochem Pharmacol 154:127–135. doi: 10.1016/j.bcp.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 485.Pitts JD, Li PC, de Wispelaere M, Yang PL. 2017. Antiviral activity of N-(4-hydroxyphenyl) retinamide (4-HPR) against Zika virus. Antiviral Res 147:124–130. doi: 10.1016/j.antiviral.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Mounce BC, Cesaro T, Moratorio G, Hooikaas PJ, Yakovleva A, Werneke SW, Smith EC, Poirier EZ, Simon-Loriere E, Prot M, Tamietti C, Vitry S, Volle R, Khou C, Frenkiel MP, Sakuntabhai A, Delpeyroux F, Pardigon N, Flamand M, Barba-Spaeth G, Lafon M, Denison MR, Albert ML, Vignuzzi M. 2016. Inhibition of polyamine biosynthesis is a broad-spectrum strategy against RNA viruses. J Virol 90:9683–9692. doi: 10.1128/JVI.01347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Cui R, Wang Y, Wang L, Li G, Lan K, Altmeyer R, Zou G. 2016. Cyclopiazonic acid, an inhibitor of calcium-dependent ATPases with antiviral activity against human respiratory syncytial virus. Antiviral Res 132:38–45. doi: 10.1016/j.antiviral.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 488.Denisova OV, Soderholm S, Virtanen S, Von Schantz C, Bychkov D, Vashchinkina E, Desloovere J, Tynell J, Ikonen N, Theisen LL, Nyman TA, Matikainen S, Kallioniemi O, Julkunen I, Muller CP, Saelens X, Verkhusha VV, Kainov DE. 2014. Akt inhibitor MK2206 prevents influenza pH1N1 virus infection in vitro. Antimicrob Agents Chemother 58:3689–3696. doi: 10.1128/AAC.02798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Badia R, Angulo G, Riveira-Munoz E, Pujantell M, Puig T, Ramirez C, Torres-Torronteras J, Marti R, Pauls E, Clotet B, Ballana E, Este JA. 2016. Inhibition of herpes simplex virus type 1 by the CDK6 inhibitor PD-0332991 (palbociclib) through the control of SAMHD1. J Antimicrob Chemother 71:387–394. doi: 10.1093/jac/dkv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Hutterer C, Eickhoff J, Milbradt J, Korn K, Zeittrager I, Bahsi H, Wagner S, Zischinsky G, Wolf A, Degenhart C, Unger A, Baumann M, Klebl B, Marschall M. 2015. A novel CDK7 inhibitor of the pyrazolotriazine class exerts broad-spectrum antiviral activity at nanomolar concentrations. Antimicrob Agents Chemother 59:2062–2071. doi: 10.1128/AAC.04534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Yu HB, Jiang H, Cheng ST, Hu ZW, Ren JH, Chen J. 2018. AGK2, a SIRT2 inhibitor, inhibits hepatitis B virus replication in vitro and in vivo. Int J Med Sci 15:1356–1364. doi: 10.7150/ijms.26125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.de Wilde AH, Li Y, van der Meer Y, Vuagniaux G, Lysek R, Fang Y, Snijder EJ, van Hemert MJ. 2013. Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J Virol 87:1454–1464. doi: 10.1128/JVI.02078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Hopkins S, Gallay P. 2012. Cyclophilin inhibitors: an emerging class of therapeutics for the treatment of chronic hepatitis C infection. Viruses 4:2558–2577. doi: 10.3390/v4112558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Cao RY, Xu YF, Zhang TH, Yang JJ, Yuan Y, Hao P, Shi Y, Zhong J, Zhong W. 2017. Pediatric drug nitazoxanide: a potential choice for control of Zika. Open Forum Infect Dis 4:ofx009. doi: 10.1093/ofid/ofx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Mueller H, Wildum S, Luangsay S, Walther J, Lopez A, Tropberger P, Ottaviani G, Lu W, Parrott NJ, Zhang JD, Schmucki R, Racek T, Hoflack JC, Kueng E, Point F, Zhou X, Steiner G, Lutgehetmann M, Rapp G, Volz T, Dandri M, Yang S, Young JAT, Javanbakht H. 2018. A novel orally available small molecule that inhibits hepatitis B virus expression. J Hepatol 68:412–420. doi: 10.1016/j.jhep.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 496.Raekiansyah M, Mori M, Nonaka K, Agoh M, Shiomi K, Matsumoto A, Morita K. 2017. Identification of novel antiviral of fungus-derived brefeldin A against dengue viruses. Trop Med Health 45:32. doi: 10.1186/s41182-017-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV Jr, Ott M. 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Musiol A, Gran S, Ehrhardt C, Ludwig S, Grewal T, Gerke V, Rescher U. 2013. Annexin A6-balanced late endosomal cholesterol controls influenza A replication and propagation. mBio 4:e00608-13. doi: 10.1128/mBio.00608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Warfield KL, Schaaf KR, DeWald LE, Spurgers KB, Wang W, Stavale E, Mendenhall M, Shilts MH, Stockwell TB, Barnard DL, Ramstedt U, Das SR. 2019. Lack of selective resistance of influenza A virus in presence of host-targeted antiviral, UV-4B. Sci Rep 9:7484. doi: 10.1038/s41598-019-43030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Nugent KM, Shanley JD. 1984. Verapamil inhibits influenza A virus replication. Arch Virol 81:163–170. doi: 10.1007/bf01309305. [DOI] [PubMed] [Google Scholar]
- 502.Schlesinger MJ, Cahill D. 1989. Verapamil and chlorpromazine inhibit the budding of Sindbis and vesicular stomatitis viruses from infected chicken embryo fibroblasts. Virology 168:187–190. doi: 10.1016/0042-6822(89)90421-2. [DOI] [PubMed] [Google Scholar]
- 503.Bajimaya S, Frankl T, Hayashi T, Takimoto T. 2017. Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virology 510:234–241. doi: 10.1016/j.virol.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Albulescu IC, Kovacikova K, Tas A, Snijder EJ, van Hemert MJ. 2017. Suramin inhibits Zika virus replication by interfering with virus attachment and release of infectious particles. Antiviral Res 143:230–236. doi: 10.1016/j.antiviral.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 505.Tan CW, Sam IC, Chong WL, Lee VS, Chan YF. 2017. Polysulfonate suramin inhibits Zika virus infection. Antiviral Res 143:186–194. doi: 10.1016/j.antiviral.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 506.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lopez MZ, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog 5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Gaither LA, Borawski J, Anderson LJ, Balabanis KA, Devay P, Joberty G, Rau C, Schirle M, Bouwmeester T, Mickanin C, Zhao S, Vickers C, Lee L, Deng G, Baryza J, Fujimoto RA, Lin K, Compton T, Wiedmann B. 2010. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology 397:43–55. doi: 10.1016/j.virol.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 508.Xia Y, Liang TJ. 2019. Development of direct-acting antiviral and host-targeting agents for treatment of hepatitis B virus infection. Gastroenterology 156:311–324. doi: 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Johari J, Kianmehr A, Mustafa MR, Abubakar S, Zandi K. 2012. Antiviral activity of baicalein and quercetin against the Japanese encephalitis virus. Int J Mol Sci 13:16785–16795. doi: 10.3390/ijms131216785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Ohnishi E, Bannai H. 1993. Quercetin potentiates TNF-induced antiviral activity. Antiviral Res 22:327–331. doi: 10.1016/0166-3542(93)90041-g. [DOI] [PubMed] [Google Scholar]
- 511.Rojas Á, Del Campo JA, Clement S, Lemasson M, García-Valdecasas M, Gil-Gómez A, Ranchal I, Bartosch B, Bautista JD, Rosenberg AR, Negro F, Romero-Gómez M. 2016. Effect of quercetin on hepatitis C virus life cycle: from viral to host targets. Sci Rep 6:31777. doi: 10.1038/srep31777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Maheshwari RK, Husain MM, Friedman RM, Krishna G. 1985. The calcium ionophore A23187 evokes and potentiates antiviral activity of interferon. J Interferon Res 5:605–612. doi: 10.1089/jir.1985.5.605. [DOI] [PubMed] [Google Scholar]
- 513.Conrad KD, Niepmann M. 2014. The role of microRNAs in hepatitis C virus RNA replication. Arch Virol 159:849–862. doi: 10.1007/s00705-013-1883-4. [DOI] [PubMed] [Google Scholar]
- 514.Onishi E, Natori K, Yamazaki S. 1991. The antiviral effect of phorbol ester and calcium ionophore A23187 is not mediated by interferons. J Interferon Res 11:171–175. doi: 10.1089/jir.1991.11.171. [DOI] [PubMed] [Google Scholar]
- 515.Yang H, Kim S-K, Kim M, Reche PA, Morehead TJ, Damon IK, Welsh RM, Reinherz EL. 2005. Antiviral chemotherapy facilitates control of poxvirus infections through inhibition of cellular signal transduction. J Clin Invest 115:379–387. doi: 10.1172/JCI23220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Marsolais D, Rosen H. 2009. Chemical modulators of sphingosine-1-phosphate receptors as barrier-oriented therapeutic molecules. Nat Rev Drug Discov 8:297–307. doi: 10.1038/nrd2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Walsh KB, Teijaro JR, Wilker PR, Jatzek A, Fremgen DM, Das SC, Watanabe T, Hatta M, Shinya K, Suresh M, Kawaoka Y, Rosen H, Oldstone MB. 2011. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc Natl Acad Sci U S A 108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, Wong G, Kobinger G, Xing Z, Couture C, Joubert P, Fritz JH, Powell WS, Divangahi M. 2014. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity 40:554–568. doi: 10.1016/j.immuni.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 519.Mehrbod P, Omar AR, Hair-Bejo M, Haghani A, Ideris A. 2014. Mechanisms of action and efficacy of statins against influenza. Biomed Res Int 2014:872370. doi: 10.1155/2014/872370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Fedson DS. 2006. Pandemic influenza: a potential role for statins in treatment and prophylaxis. Clin Infect Dis 43:199–205. doi: 10.1086/505116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, Seeger W, Lohmeyer J. 2008. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med 205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Aldridge JR Jr, Moseley CE, Boltz DA, Negovetich NJ, Reynolds C, Franks J, Brown SA, Doherty PC, Webster RG, Thomas PG. 2009. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci U S A 106:5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Moseley CE, Webster RG, Aldridge JR. 2010. Peroxisome proliferator-activated receptor and AMP-activated protein kinase agonists protect against lethal influenza virus challenge in mice. Influenza Other Respir Viruses 4:307–311. doi: 10.1111/j.1750-2659.2010.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Cloutier A, Marois I, Cloutier D, Verreault C, Cantin AM, Richter MV. 2012. The prostanoid 15-deoxy-Delta12,14-prostaglandin-j2 reduces lung inflammation and protects mice against lethal influenza infection. J Infect Dis 205:621–630. doi: 10.1093/infdis/jir804. [DOI] [PubMed] [Google Scholar]
- 525.Qing M, Zou G, Wang QY, Xu HY, Dong H, Yuan Z, Shi PY. 2010. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob Agents Chemother 54:3686–3695. doi: 10.1128/AAC.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]