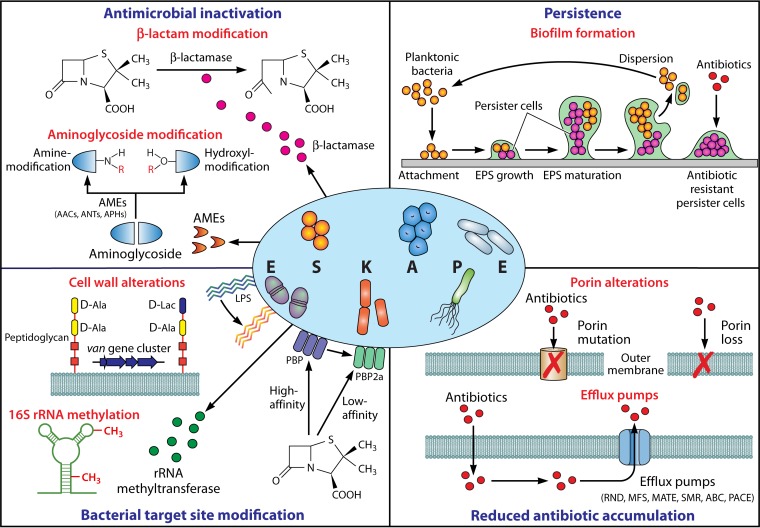
FIG 1.
Mediators of ESKAPE pathogen antimicrobial resistance. Mechanisms facilitating antimicrobial resistance in ESKAPE pathogens can be broadly categorized into four groups: (i) enzyme-mediated antimicrobial inactivation, which either irreversibly destroys the active antibiotic site (e.g., hydrolytic cleavage of the β-lactam ring by β-lactamases) or covalently modifies key structural elements of the drug to hinder the bacterial target site interaction (e.g., aminoglycoside-modifying enzymes that catalyze hydroxyl/amino group modifications); (ii) bacterial target site modification, which prevents the binding or which reduces the affinity of the antibiotic molecule at the cell surface (e.g., LPS modification, PBP2a expression with reduced β-lactam affinity, and van gene cluster-mediated peptidoglycan modification) or intracellularly (e.g., 16S RNA methylation); (iii) reduced antibiotic accumulation through the mutation or loss of outer membrane channels (e.g., OprD in P. aeruginosa, CarO in A. baumannii, and OmpK36 in K. pneumoniae) and expression of efflux systems to actively extrude drugs out of the cell (e.g., RND, MFS, MATE, SMR, ABC, and PACE); and (iv) persistence through biofilm-embedded cells which demonstrate a markedly higher tolerance to antimicrobial agents than planktonic bacteria. AMEs, aminoglycoside-modifying enzymes; AACs, aminoglycoside acetyltransferases; ANTs, aminoglycoside nucleotidyltransferases; APHs, aminoglycoside phosphotransferases; LPS, lipopolysaccharide; PBP, penicillin-binding protein; RND, resistance-nodulation-division; MFS, major facilitator superfamily; MATE, multidrug and toxic compound extrusion; SMR, small multidrug resistance; ABC, ATP-binding cassette; PACE, proteobacterial antimicrobial compound efflux; EPS, extracellular polymeric substance.