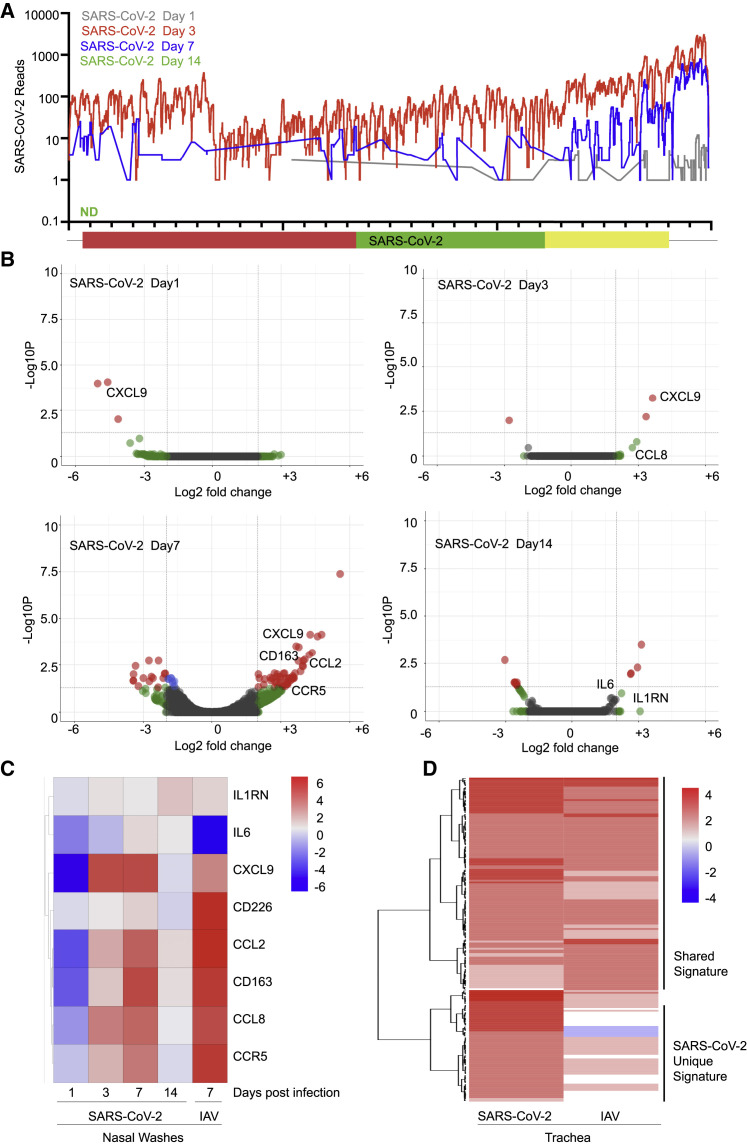
Figure 3.
Longitudinal Analysis of the Host Response to SARS-CoV-2 in Ferrets
(A) Read coverage along the SARS-CoV-2 genome. The graph indicates the number of viral reads per each position of the virus genome identified in RNA extracted from nasal washes of ferrets 1 (gray), 3 (red), 7 (blue), and 14 (green) days after infection (ND, not detected).
(B) Volcano plots indicating DEGs of ferrets along the course of a SARS-CoV-2 infection as in (A). DEGs (p-adjusted < 0.05) with a |log2(fold change)| of more than 2 are indicated in red. Non-significant DEGs with a |log2(fold change)| of more than 2 are indicated in green.
(C) Heatmap depicting the expression levels of a subset of cytokines differentially expressed in nasal washes collected from ferrets infected with the indicated viruses at specific times.
(D) Heatmap depicting the expression levels of lymphoblast-related genes differentially expressed in trachea samples collected from ferrets infected with the indicated viruses after 3 days. The graphs show the log2(fold change) of DEGs of infected compared with mock-infected animals. Genes included have a log2(fold change) of more than 2 and a p-adjusted value of less than 0.05. Ferrets were randomly assigned to the different treatment groups (naive, n = 2; SARS-CoV-2 infection, n = 6; IAV [pH1N1] infection, n = 2; IAV [H3N2] infection, n = 2).