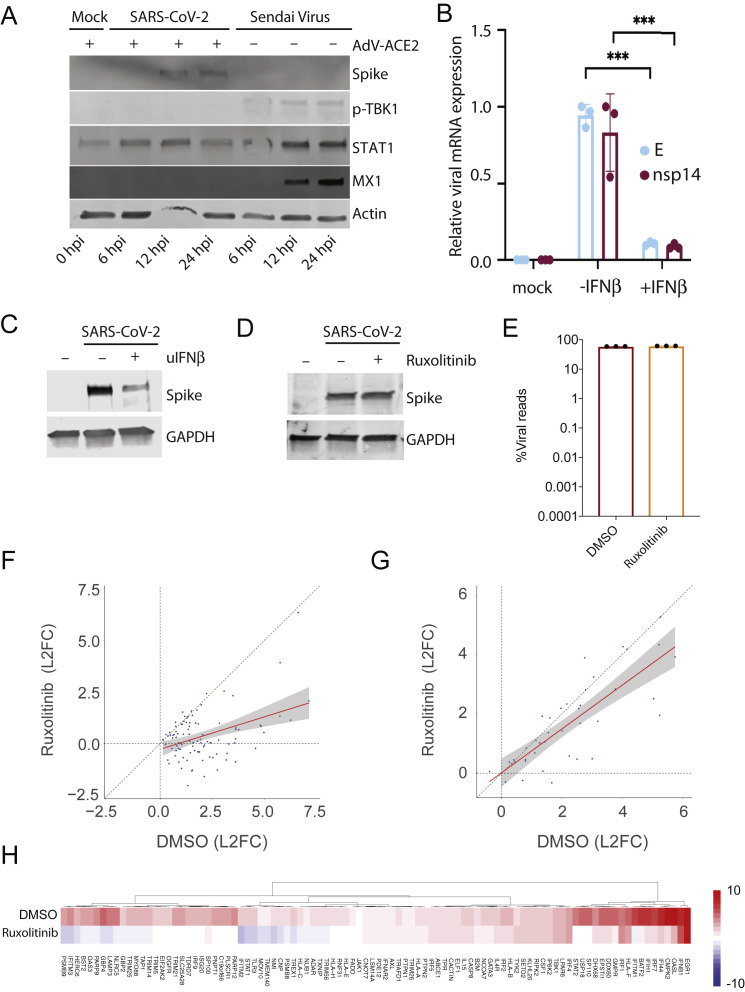
Figure S1.
Role of IFN Response in Infection with SARS-CoV-2, Related to Figure 1
(A) Western blot analysis of WT or ACE2-expressing A549 cells mock-treated or infected with SARS-CoV-2 or Sendai virus. Whole cell lysates were analyzed by SDS-PAGE and blotted for SARS-CoV-2 spike, phospho-TBK1, MX1, STAT1 and actin. (B) qRT-PCR analysis of Vero E6 cells infected with SARS-CoV-2 and treated with IFNβ 2 h post infection as indicated. The graph depicts the relative amount of SARS-CoV-2 envelope (E) and non-structural protein 14 (nsp14) normalized to human α-Tubulin. Error bars represent the standard deviation of the mean fold change of three independent biological replicates. Statistical significance calculated by Student's t test corrected for multiple comparisons using Holm-Sidak method (∗∗∗) p value < 0.001. (C) Western blot analysis of conditions as in (B). Whole cell lysates were analyzed by SDS-PAGE and blotted for SARS-CoV-2 spike and GAPDH. (D) Western blot analysis of ACE2-expressing A549 cells infected with SARS-CoV-2 with or without Ruxolitinib. Whole cell lysates were analyzed by SDS-PAGE and blotted for SARS-CoV-2 spike and GAPDH. (E) Virus replication levels in SARS-CoV-2-infected A549-ACE2 cells treated with or without Ruxolitinib. RNA-seq was performed on polyA enriched total RNA and the percentage of virus-aligned reads (over total reads) is indicated for each sample. Error bars represent standard deviation from three independent biological replicates. Infections were preformed at high MOI (MOI: 2). (F-G) Expression levels of (F) ISGs or (G) cytokines and chemokines in conditions as in (E). Scatterplot of the log2(Fold Change) of individual genes in SARS-CoV-2-infected A549-ACE2 cells treated with or without Ruxolitinib. Linear regression line and confidence interval (95%) is shown as a red line and gray shaded area, respectively. Dotted diagonal represent no changes between conditions. (H). Heatmap depicting the expression levels of ISGs as in (F).