Figure 2.
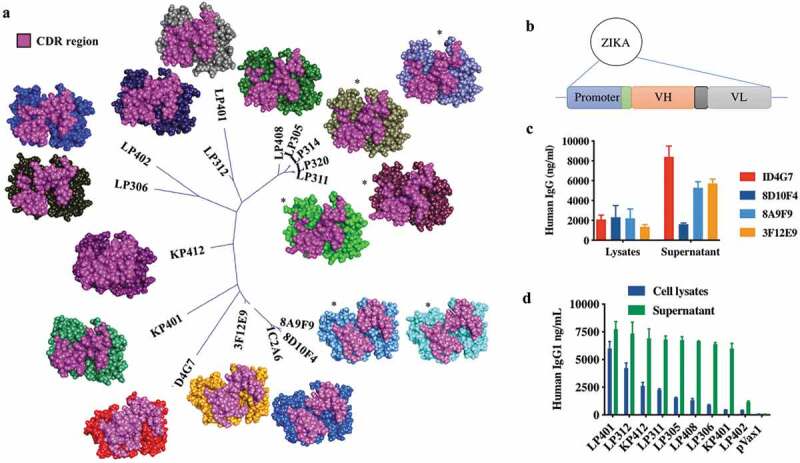
Comparison of Zika envelope antibodies by CDR phylogeny and molecular modeling.
(a) An unrooted phylogenetic tree based on CDR comparisons (only) of 16 anti-Zika envelope antibodies was generated to demonstrate relatedness and by extension the potential for epitope targeting similarity. Shown nearby to each antibody location on the tree is a molecular model generated with standard antibody modeling protocols as implemented in Discovery Studio. Models are shown in CPK format and CDRs are highlighted. As sequence relatedness translates to structural similarities, it can be noted that clusters of nearly or completely identical CDRs adopt similar conformations. Of the Fv models shown, LP408, LP314, and 8A9F9 have been subjected to docking analysis using ZDOCK as implemented in Discovery Studio and scored with ZRANK. In all cases, the EDIII region (a known target for effective neutralizing antibodies) contains multiple top poses for the antibodies in question. In particular, LP408 and LP314 demonstrate the top 15 of 20 poses total in this region. * indicates similar CDR conformations between closely related mAbs and a potential ZIKV ENV Domain III (EDIII) binding for clones 8A9F9, LP314, and LP408. (b) Schematic diagram of ZIKV-specific antibody heavy and light chain encoded as a codon-optimized single cassette into the pVax1 DNA plasmid. Construction and expression of ZIKV-dMAb plasmid constructs from (c) ZIKV DNA plasmid-vaccinated murine spleen-derived and (d) ZIKV-infected RhMac spleen-derived. Each dMAb construct was transfected into 293T cells in order to determine in vitro expression via enzyme-linked immunosorbent assays (ELISAs). The bar represents the level of IgG present at 48-h post transfection for both cell lysates and supernatants per construct.
