Table 4.
Chemical structures and binding affinities of analogues 9a-s at native iGluRs (rat synaptosomes) and homomeric recombinant iGluRs. All values in μM.a
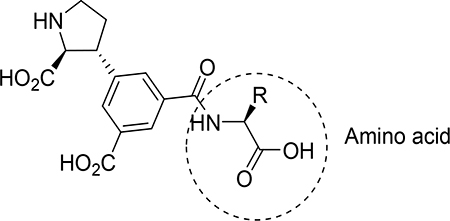 | |||||||
|---|---|---|---|---|---|---|---|
| Native iGluRs (synaptosomes)b | Cloned homomeric iGluRsc | ||||||
| Amino acid | AMPA IC50 | KA IC50 | NMDA Ki | GluK1 Ki | GluK2 Ki | GluK3 Ki | |
| 8a | Gly | 17 [4.76 ± 0.04] | 44 [4.37 ± 0.06] | 0.60 [6.23 ± 0.05] | 25.2 ± 1.8 | ’100 | 22.3 ± 6.0 |
| 8b | L- Ala | 32 [4.52 ± 0.10] | 54 [4.28 ± 0.06] | 0.87 [6.06 ± 0.02] | 13.3 ± 0.5 | ’100 | 17.8 ± 2.1 |
| 8c | L- Val | 27 [4.57 ± 0.02] | 27 [4.57 ± 0.05] | 3.4 [5.47 ± 0.04] | 4.41 ± 0.63 | ’100 | 5.80 ± 0.54 |
| 8d | L- Leu | 33 [4.49 ± 0.07] | 66 [4.20 ± 0.09] | 3.9 [5.41 ± 0.03] | 11.7 ± 1.4 | ’100 | 20.6 ± 3.1 |
| 8e | L- Ile | 10–50 | 10–50 | 5–10 | (68 ± 5)d | (91 ± 6)d | (51 ± 6)d |
| 8f | L- Phe | 10–50 | >50 | 1–5 | (86 ± 8)d | (103 ± 10)d | (66 ± 7)d |
| 8g | L-Tyr | 10 | 10–50 | 0.96 [6.02 ± 0.01] | (62 ± 1)d | (84 ± 9)d | 5.69 ± 0.90 (40 ± 7)d |
| 8h | L- Ser | 10–50 | >50 | 1–5 | (75 ± 2)d | (89 ± 14)d | (63 ± 6)d |
| 8i | L- Thr | 10–50 | 10–50 | 5 | (80 ± 2)d | (104 ± 7)d | (55 ± 10)d |
| 8j | L- Pro | 10–50 | 10–50 | 1.1 [5.98 ± 0.03] | 1.44 ± 0.18 (35 ± 1)d | (86 ± 1)d | 2.41 ± 1.06 (18 ± 4)d |
| 8k | L- Glu | 10–50 | >100 | 0.83 [6.08 ± 0.01] | (91 ± 6)d | (100 ± 14)d | (86 ± 3)d |
| 8l | L- Gln | -- | -- | -- | -- | -- | -- |
| 8m | L- Asp | 5–10 | >100 | 1–5 | (86 ± 7)d | (102 ± 5)d | (83 ± 6)d |
| 8n | L- Asn | -- | -- | -- | -- | -- | -- |
| 8o | L- Lys | 10–50 | 10–50 | 10–50 | 3.27 ± 0.55 (50 ± 5)d | (79 ± 4)d | 5.06 ± 0.03 (37 ± 2)d |
| 8p | L- Cys | 10–50 | 10–50 | 1–5 | (80 ± 2)d | (88 ± 6)d | 10–50 (60 ± 4)d |
| 8q | L- His | 10–50 | 10–50 | 5 | (76 ± 1)d | (66 ± 1)d | 6.23 ± 2.69 (44 ± 3)d |
| 8r | L- Trp | 10 | 10–50 | 1–5 | (72 ± 3)d | (90 ± 6)d | 10–50 (62 ± 5)d |
| 8s | L- Arg | 10 | 10 | 10–50 | 2.65 ± 1.06 (40 ± 1)d | 10–50 (66 ± 1)d | 2.78 ± 1.41 (29 ± 9)d |
--: Analog unstable in buffer - values not determined. Radio ligands used: AMPA, [3H]AMPA; KA, [3H]KA; [3H]CGP-39653; GluK1, [3H]SYM2081; GluK2 and GluK3, [3H]KA
Data are mean values of three to six individual experiments performed in triplicate. For AMPA and KA: pIC50 values with SEM in brackets. For NMDA: pKi values with SEM in brackets.
Mean values ± SEM of at least three experiments conducted in triplicate at 12–16 drug concentrations.
%specific binding mean values ± SD of three experiments performed at 10 μM ligand concentration.
