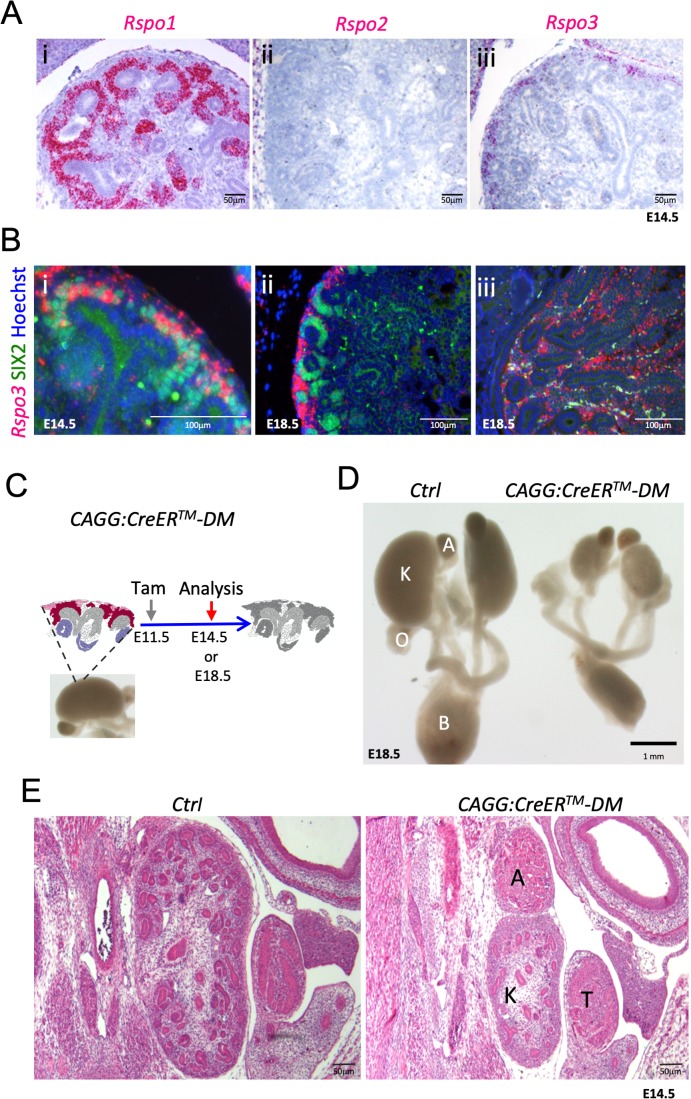
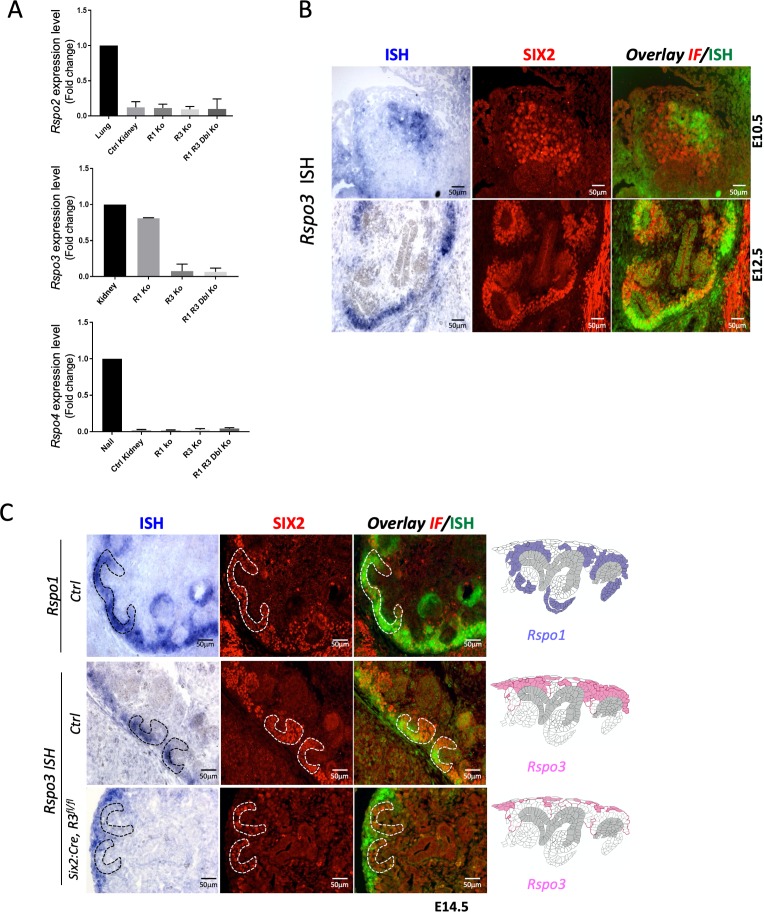
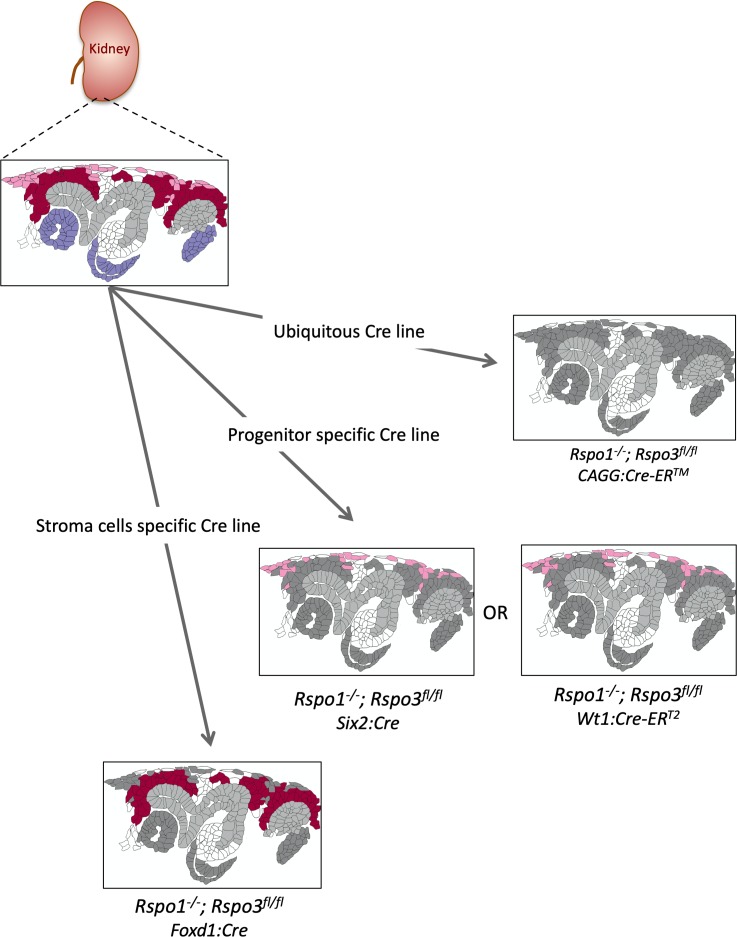
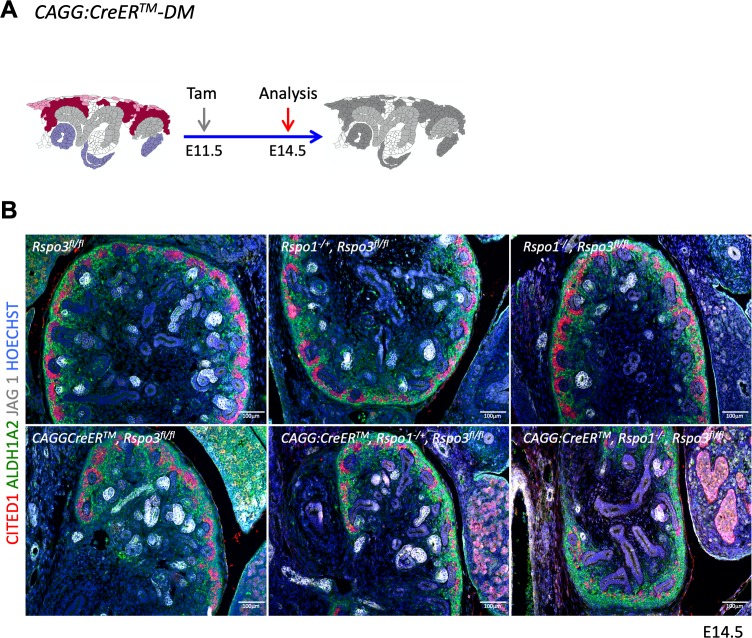
Figure 1. Rspo1 and Rspo3 are expressed in embryonic kidneys and are required for normal development.
(A) RNA-Scope analysis demonstrates Rspo1 (i) and Rspo3 (iii) expression in the nephrogenic zone of developing (E14.5) kidneys. (B) RNA-Scope analysis followed by immunostaining for the progenitor marker SIX2 reveals a switch from strong Rspo3 expression within progenitors at E14.5 (i) to almost exclusively stromal progenitor expression at E18.5 (ii). In addition, strong staining was found within medullary stromal cells (iii). Hoechst stains nuclei in blue (C) Schematic outline of tamoxifen induction for CAGG:CreERTM-mediated deletion leading to a complete loss of R-spondin expression in the Rspo1/Rspo3 double mutant (DM). Colour legend: Purple for Rspo1+, pink for Rspo3+, dark red for Rspo1/Rspo3+ cells. Rspo depleted cells are dark grey, light grey highlights ureteric cells. (D) Macroscopic view of the urogenital system of Control and CAGG:CreERTM-DM embryos dissected at E18.5, (E). Hematoxylin and Eosin (Hand E) staining of E14.5 sections reveals smaller kidneys virtually lacking nephrons. A: adrenal gland, B: bladder, K: kidney, O: ovary, T: testis. CAGG:CreERTM-DM stands for (CAGG:CreERTM, Rspo1-/-, Rspo3fl/fl), Ctrl: Control.