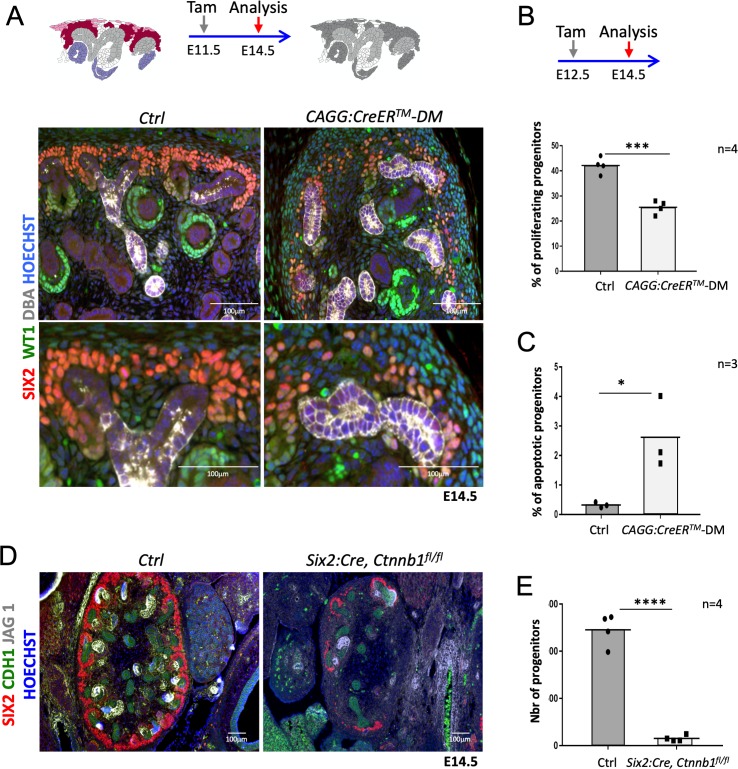
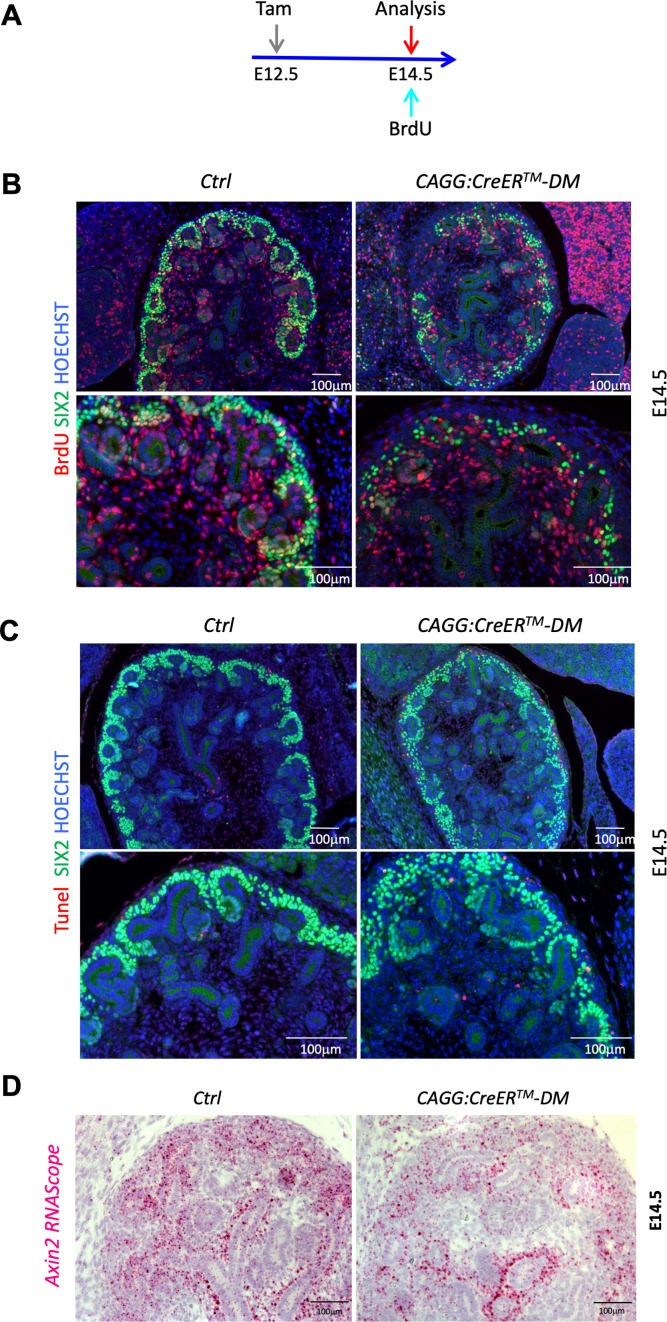
Figure 2. R-spondins are required for renal progenitor maintenance.
(A) Immunofluorescent analysis at E14.5 (induced at E11.5) reveals loss of SIX2+ progenitor cells and nascent nephrons (comma or S-shaped bodies) in CAGG:CreERTM-DM embryos. (WT1 = green; SIX2 = red; DBA = white; Hoechst = blue). Colour legend for the cartoon: Purple label Rspo1+, pink Rspo3+, dark red Rspo1/Rspo3+ cells. Rspo depleted cells are dark grey, light grey highlights ureteric cells. (B) Quantification of BrdU-labelled SIX2+ progenitors performed on four embryos (n = 4) demonstrates a significant reduction of proliferation 2 days after Rspo3 deletion. See Figure 2—source data 1 (C) TUNEL analysis reveals a dramatic increase in apoptosis (n = 3 embryos for each genotype, two litters). (D) Progenitor specific deletion of β-catenin (Six2:Cre; Ctnnb1fl/fl) results in the loss of progenitor cells at E14.5 (SIX2 = red; CDH1 = green; JAG1 = white). (E). Quantification of SIX2+ progenitors (n = 4 embryos for each genotype isolated from two litters). See Figure 2—source data 1. Columns are means ± SEM with p<0.05 (*), p<0.01 (**), p<0.001 (***), p<0.0001 (****). One black dot = average value for one control embryo, one black square = average value for one CAGG:CreERTM-DM embryo.