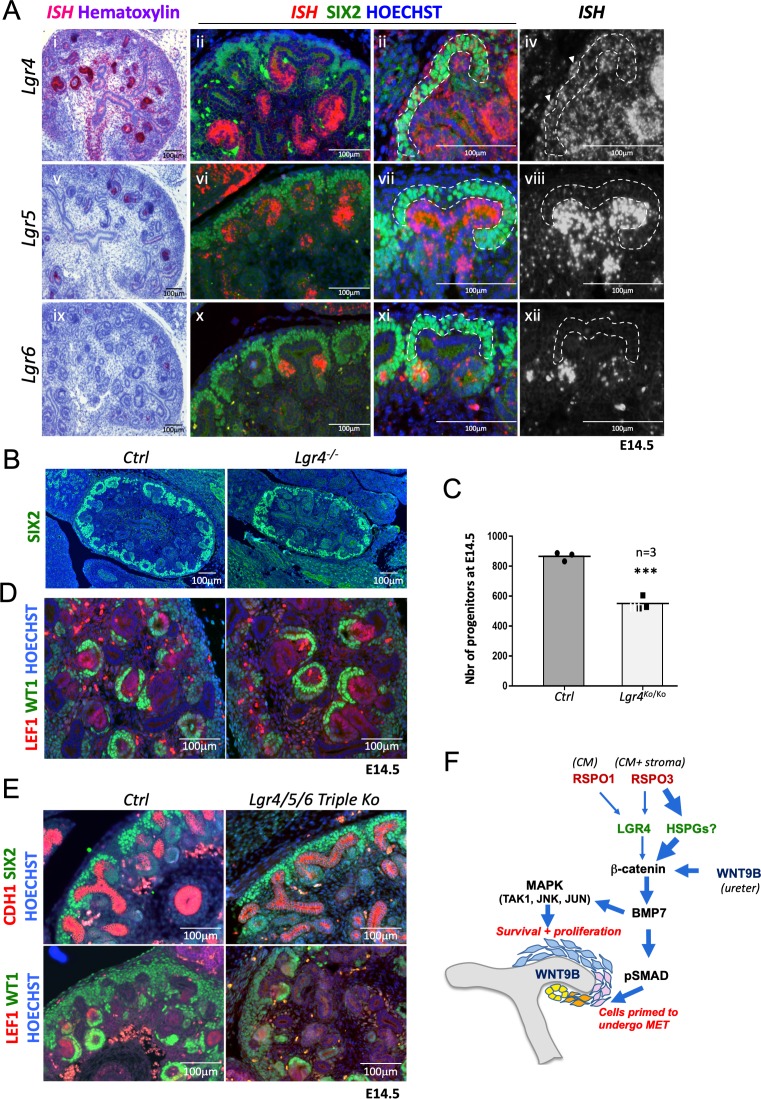
Figure 6. R-spondins can function in an LGR-independent manner during kidney development.
(A) i-iiii) RNAScope analysis (red) revealed low levels of Lgr4 expression throughout the developing kidney with strong signal within the distal portion of the forming nephron and weak activation in the stromal cells (white arrowhead). v-viii) Lgr5 expression was found within the ureteric tip and distal segment of S-shaped bodies. ix-xii) Lgr6 expression was restricted to PTAs of newly forming nephrons. (SIX2 = green, Hoechst = blue). The Cap Mesenchyme compartment is outlined by dotted white lines. (B) Immunofluorescence analysis of Lgr4 knockout and control samples performed on E14.5 kidney sections with SIX2 antibodies reveals a reduction of nephron progenitors. (C) Quantification of SIX2+ progenitors from (B) (for each genotype n = 3 embryos collected from two litters,) Every black dot or square represents the total number of SIX2+ progenitors located in the CM counted in one control or mutant of an entire kidney field, see Figure 6—source data 1. (D) Lgr4 negative progenitors undergo MET as revealed by WT1 staining (high WT1 expression is found in the proximal part of Comma and S-shaped bodies, as well as podocytes). (E) Immunofluorescent analysis in wholebody Lgr4/5/6 mutants demonstrates persistence of progenitors and MET despite the absence of all three cognate R-spondin receptors. (F) Model for the molecular cascade regulated by R-spondins during nephrogenesis.