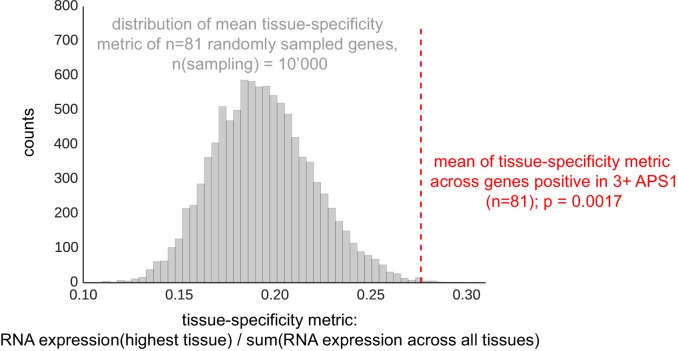
Figure 2. PhIP-Seq identifies novel (and known) antigens across multiple APS1 sera.
(A) Hierarchically clustered (Pearson) z-scored heatmap of all genes with 10-fold or greater signal over mock-IP in at least 3/39 APS1 sera and in 0/28 non-APS1 sera. Black labeled antigens (n = 69) are potentially novel and grey labeled antigens (n = 12) are previously literature-reported antigens.