Figure 1. Epigenomic Characteristics of Young Rod Photoreceptors.
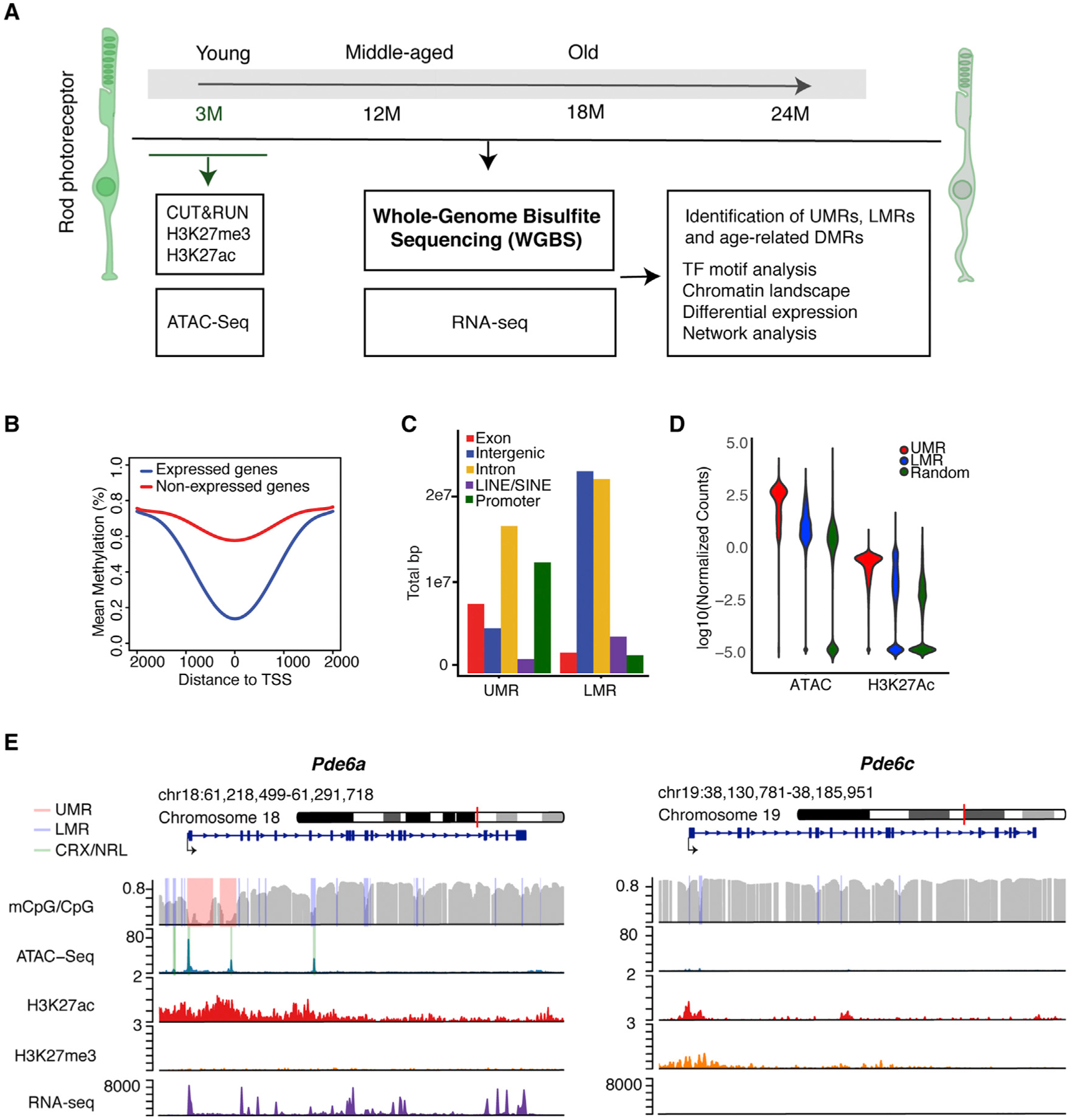
(A) Outline of the experimental paradigm. Integrative analysis of base-resolution DNA methylation and RNA-seq profiles from young (3-month-old), middle-aged (12-month-old), and old (18-and 24-month-old) rods with chromatin accessibility and histone marks from 3-month-old rods.
(B) DNA methylation levels around the TSS of expressed and unexpressed genes in 3-month-old rods.
(C) Distribution of UMRs and LMRs across various genomic regions of 3-month-old rods.
(D) Normalized ATAC-seq and H3K27ac read numbers over total region size of UMRs, LMRs, and random genomic regions of similar size in 3-month-old rods.
(E) Examples of UMRs (pink) and LMRs (purple) in Pde6a and Pde6c. CRX and NRL ChIP-seq peaks are shown in green. ATAC-seq peaks overlap with CRX and NRL binding in both UMRs and LMRs within the rod-specific Pde6a gene. CpG methylation, ATAC-seq, H3K27ac, H3K27me3, and RNA-seq profiles are shown for the 3-month time point over the entire gene and its surroundings.
DMR, differentially methylated region; LMR, low methylated region; TF, transcription factor; TSS, transcription start site; UMR, unmethylated region.
