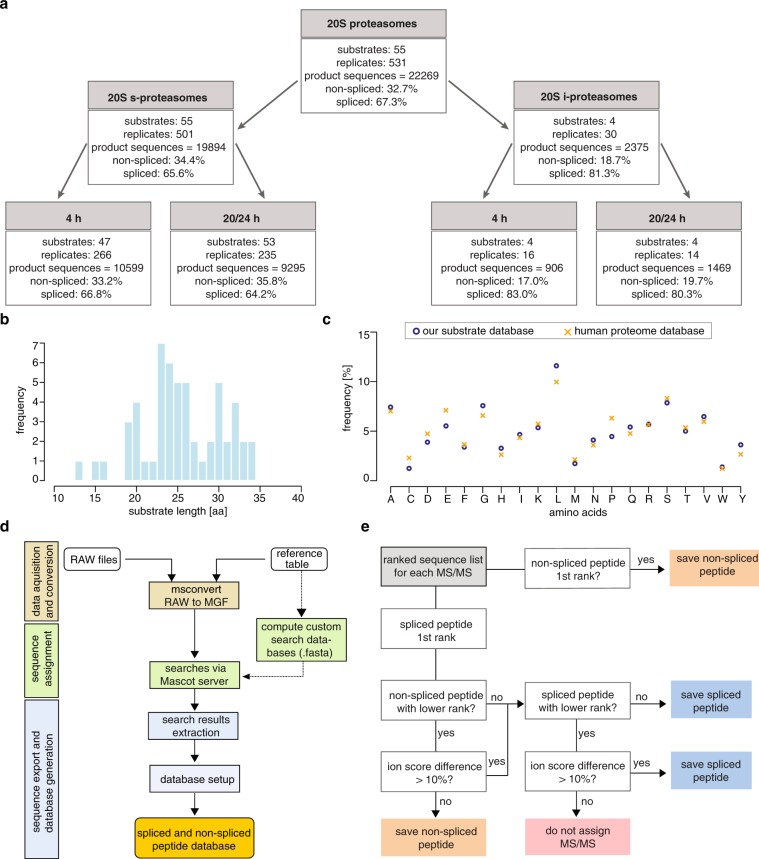
Fig. 2.
Database content and construction. (a) Database content. The database consists of peptide products identified in digestions of 55 unique substrates with 20 S and 26S s- and i-proteasomes at 4 h and 20/24 h digestion time. Shown is the main part, which represents all digestions with 20 S proteasomes. Values refer to product sequences (identified per sample). Several product sequences are identified in multiple experimental conditions using the sample substrate. Therefore, the number of unique peptide sequences, i.e. peptide sequences identified per substrate, is smaller than that of product sequences. (b) Length distribution of synthetic polypeptide substrates included in the database. (c) Matrixes of the amino acid frequency of the synthetic polypeptide substrates included in our database and of the human proteome database. (d) Overview of the data processing pipeline to construct the database. (e) Identification of spliced and non-spliced peptides from MS datasets, which was applied to assign peptides to each MS/MS spectrum.