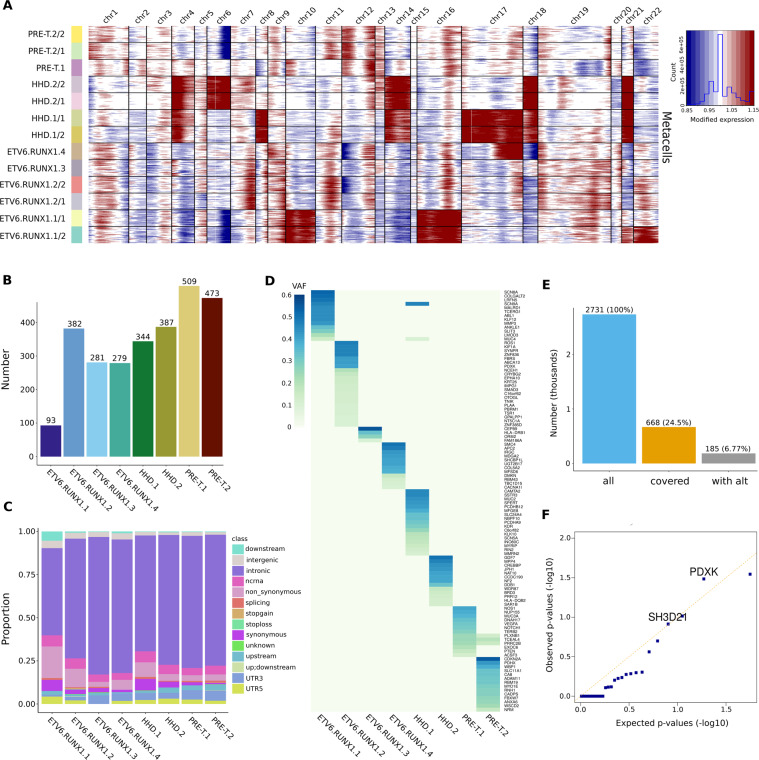
Figure 4.
Somatic alterations and intra-individual expression variability. (A) Copy number profiles of samples and intra-individual transcriptional clusters using equal size metacells and healthy pediatric BMMCs as control cells. (B) Number of somatic mutations in cALL samples. (C) Genomic annotations of somatic mutations in cALL samples. (D) Gene names and variant allele frequencies of exonic non-synonymous somatic mutations in cALL samples. (E) Number of somatic mutations used as input to obtain allele calls from single cell RNA-seq alignment files using vartrix (all = number of GRCh38 mutations lifted from hg19; covered = number of mutations covered in at least one cell; with alt = number of mutations with at least one mutant allele call). (F) QQ-plot of Fisher’s exact test p-values of somatic mutations tested for enrichment in intra-individual transcriptional clusters (>0.1% of cells with mutant allele call, n = 31 mutations).