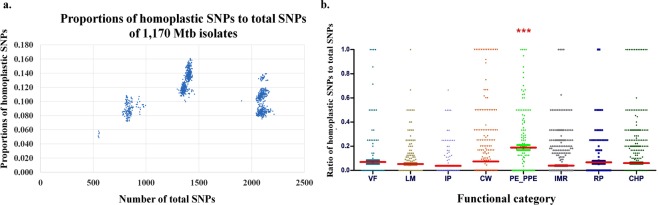
Figure 1.
(a) The plots between the ratios of the numbers of homoplastic SNPs per total SNPs and the total SNPs of the 1,170 Mtb isolates. The ratios of homoplastic SNPs per total SNPs were markedly different between lineages so that the major groups from left to right are L4 (0.089 ± 0.011), L2 (0.130 ± 0.013), L3 (0.110 ± 0.006), and L1 (0.099 ± 0.015), respectively. The small leftmost group represents L4.8 which is most closely related to H37Rv, a member of L4.9. (b) The ratio of the number of homoplastic SNPs to total SNPs of each Mtb gene categorized into 8 functional categories: i) virulence and detoxification (VF, 226 genes), ii) lipid metabolism (LM, 238 genes), iii) information pathway (IP, 232 genes), iv) cell wall and cell process (CW, 751 genes), v) pe/ppe family protein (PE_PPE, 168 genes), vi) intermediary metabolism and respiration (IMR, 898 genes), vii) regulatory protein (RP, 193 genes), viii) conserved hypothetical protein (CHP, 1,163 genes). Each dot represents a gene that harbored at least a SNP. Statistical differences were evaluated with the nonparametric Kruskal-Wallis test. Asterisks (***) indicate the statistical significance (p < 0.001).