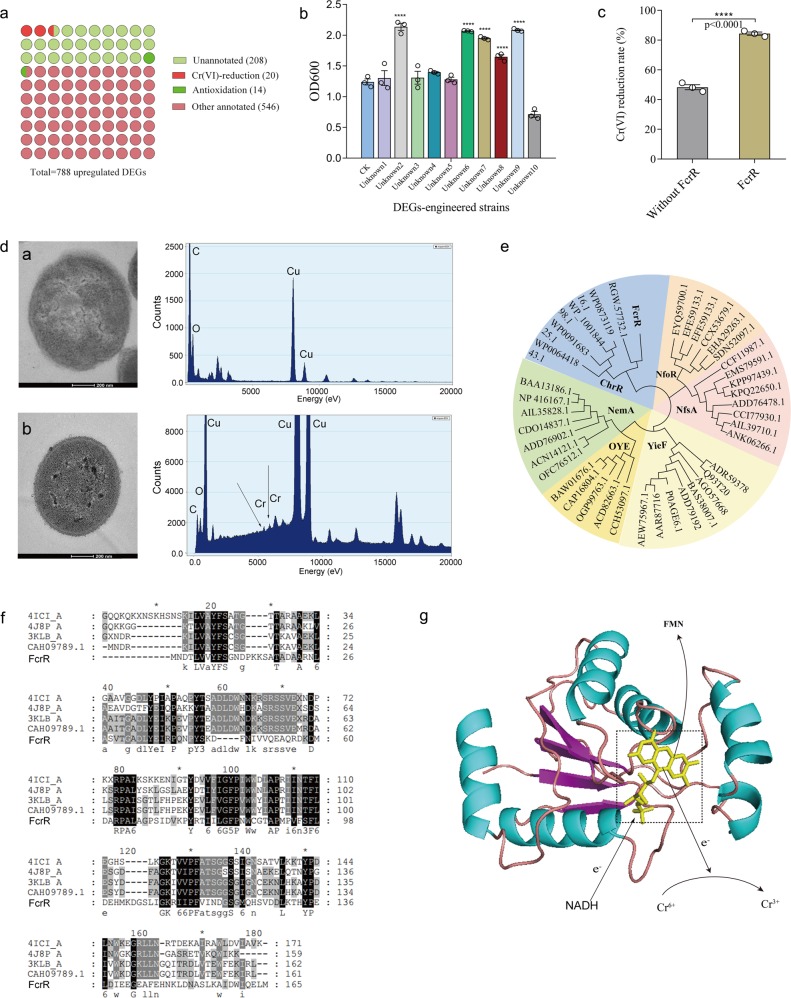
Fig. 5. Functional analysis of selected unannotated DEGs.
a The distribution of Cr remediation relevant genes in total DEGs. b The growth of engineered E. coli strains harboring DEGs in LB medium with 1 mM Cr(VI) (n = 3). CK represents E. coli BL21 containing the empty plasmid pET28a (+), unknown 1–10 represents E. coli BL21 with pET28a harboring gene comp53346, 53095, 24337, 53087, 53032, 51796, 52516, 53047, 53431, and 53299, respectively. c Cr(VI) reduction rate of engineered E. coli BL21 with 53431 (n = 3). d TEM and EDX analyses of E. coli FcrR after inoculated in LB medium and incubated overnight without Cr(VI) or with 1 mM Cr(VI), and the corresponding EDX spectra. Arrows show intracellular Cr(III) precipitates, and scale bars: 200 nm. e Evolutionary relationship of FcrR with known chromate reductases from different genera. A composite evolutionary tree with the FcrR was generated using the Neighbor-Joining method based on the reported chromate reductase families (NfoR, NfsA, YieF, OYE, NemA, and ChrR). f Sequence analysis of FcrR homologs. The highly conserved residues are highlighted in black, and other residues with high levels of similarity are highlighted in gray. g Structural modeling of FcrR. For b, data significance was analyzed using the one-way ANOVA followed by Tukey’s post hoc test. For c, data significance was analyzed using student’s t-test. Significance was marked as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.