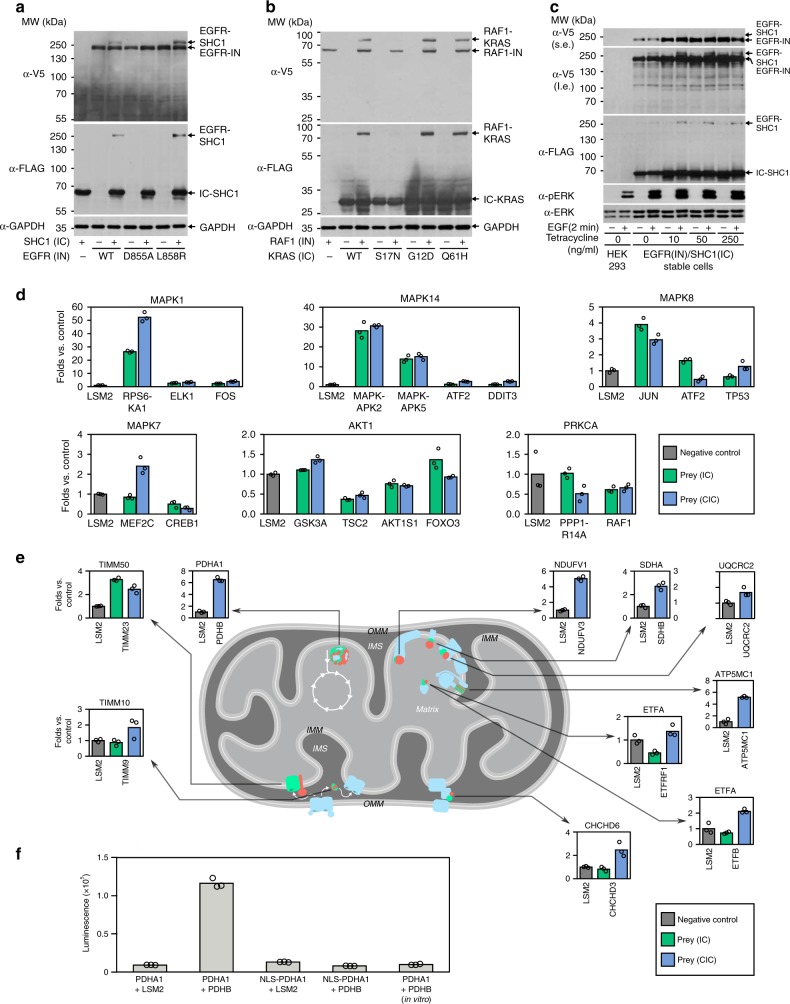
Fig. 4. Detection of physiological PPIs and their inhibition with SIMPL.
a EGFR/SHC1 interaction. EGFR WT, inactive (D855A), or constitutively active (L858R) mutants in IN format were co-expressed with SHC1 (IC) in HEK 293 cells. Their interactions were analyzed with western analysis. b KRAS/RAF interaction. RAF1-IN and IC-KRAS (WT, inactive S17N mutant, or active G12D or Q61H mutant) were transiently expressed in HEK 293 cells followed by western analysis. c Stable cells derived from HEK 293 T-Rex FlpIn with EGFR (IN) and SHC1 (IC) inserted into the FRT site were treated with the indicated different concentrations of tetracycline for 6 h, followed by treatment with EGF (100 ng/ml) for 2 min and then analysis by western blot. s.e. short exposure, l.e. long exposure. Each blot in a–c is representative of three independent experiments. d SIMPL analysis of kinase/substrate interactions. The indicated kinase-IN constructs were individually expressed along with their substrates in either IC or CIC format and their interactions were detected by ELISA assay. LSM2 was used as a negative control prey. e Mitochondrial PPIs. The selected mitochondrial bait proteins were constructed in IN format. They were then co-expressed with the indicated preys in either IC or CIC format, or in both. The interactions were examined with ELISA. LSM2 was used as a negative control prey. OMM outer mitochondrial membrane, IMS intermembrane space, IMM inner mitochondrial membrane. f Retest of PDHA1/PDHB interaction with ELISA-coupled SIMPL. In one sample (the fourth from left), the mitochondrial targeting sequence of PDHA1 was replaced by a nuclear localization sequence. In the last sample (the first from right), WT PDHA1 and PDHB were expressed in separate cells and the cell lysates were mixed for ELISA. The experiments in d–f were performed in triplicates and their mean values are presented as bars with each replicate shown in a single dot. Source data are available in the Source Data file.