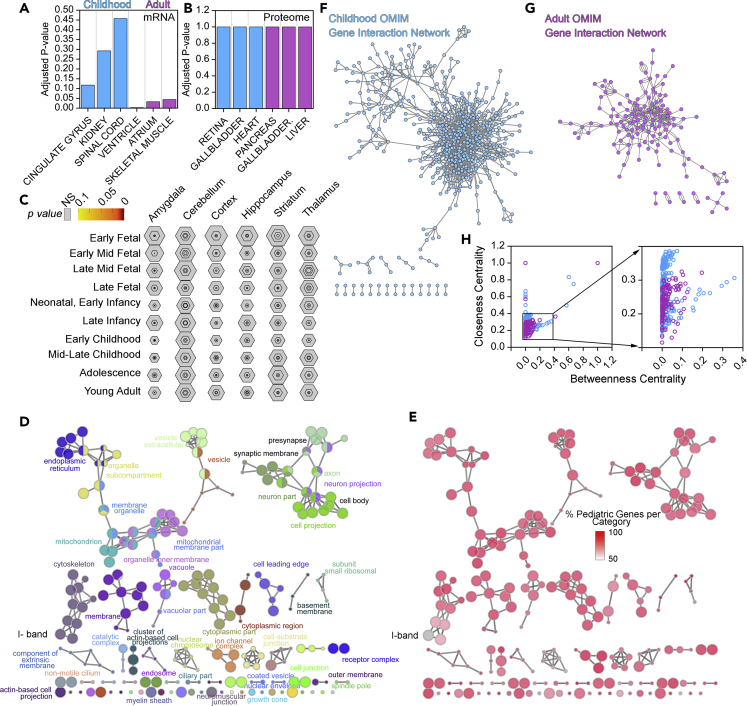
Figure 4.
Rare Disease Gene Ontologies and Protein-Protein Interaction Topologies
(A–C) We used the childhood and adult gene HPO lists to explore tissue expression in human tissues using the ARCHS4 transcript database (A), the Human Proteome Map Database (B), or de human brain developmental mRNA expression database CSEA (C) (Kim et al., 2014, Lachmann et al., 2018, Wells et al., 2015). A and B present top ranked tissues where gene lists are expressed. Note that only adult disease mRNAs are significantly enriched in categories describing striated muscle. Childhood diseases genes do not enrich nervous tissue or any other tissue ontologies. Figures A–C show Fisher's exact p values, followed by the Benjamini-Hochberg correction.
(D and E) Gene lists for childhood and adult diseases were combined and analyzed using the Cytoscape ClueGo plugin for cellular compartment ontologies (GO:CC) (Bindea et al., 2009). Color in D depicts GO CC terms. Color in E represents the percentage of genes that belong to childhood diseases in the CC term depicted in (D). Note that only one term is equally represented by childhood and adult genes: the I- band belonging to striated muscle. All ontologies are significant with a corrected p value <0.05. Size of circle is proportional to the significance of the term.
(F–H) Protein-protein interaction network data for the childhood (F) and adult disease gene lists (G) were obtained from Genemania (Franz et al., 2018). Networks were built and their topologies analyzed with Cytoscape and the NetworkAnalyzer plugin (Doncheva et al., 2012, Shannon et al., 2003). H presents centrality parameters for the childhood (blue symbols) and adult disease genes (purple symbols).