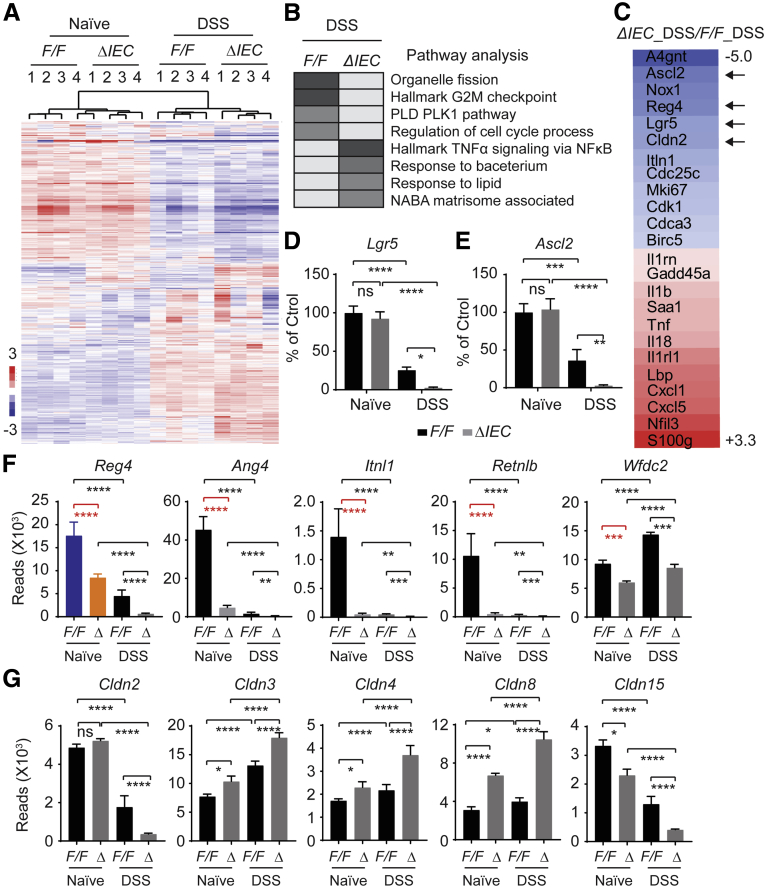
Figure 3.
RNA sequencing and pathway analysis.NCoR1F/F and NCoR1ΔIEC mice were treated with water or DSS. On day 3, colon RNA was prepared. Samples from 3 mice were combined as 1 sample, with 4 samples from each group subjected for RNA sequencing analysis. Total RNA was used for preparation of the sequencing libraries using the Illumina Trueseq RNA Sample Prep Kit, and the sequencing was performed on a Hiseq4000 (Illumina, San Diego, CA). RNA-seq reads were aligned to the mouse mm10 genome. (A) Heat map comparing gene expression differences between NCoR1F/F and NCoR1ΔIEC mice treated with either water or DSS. (B) Pathway analysis of DSS-NCoR1F/F vs DSS-NCoR1ΔIEC mice. (C) Representative markers of down-regulated (in blue) and up-regulated (in red) genes between DSS treated NCoR1F/F and NCoR1ΔIEC mice. (D and E) RNA sequencing reads of intestinal stem cell marker genes Lgr5 and Ascl2. The data are presented as the percentage change when compared with naïve NCoR1F/F mice. (F) Transcriptional alteration of DCS cell marker Reg4 and AMP genes. (G) Expression of the Cldns. The data are described as reads in fragments per kilobase of exon per million mapped reads. Ctrol, control; PLD, Phospholipase D; PLK, polo-like kinase-1. *P < .05, **P < .01, ***P < .001, and ****P < .0001.