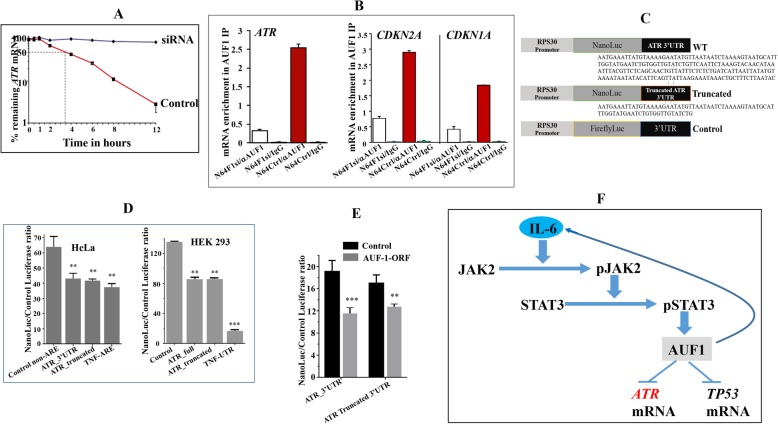
Fig. 2.
AUF1 destabilizes the ATR mRNA through binding its 3′UTR. a TCF-64 cells bearing either a scrambled sequence (control) or AUF1siRNA (siRNA) were treated with actinomycin D for the indicated periods of time. Total RNA was extracted and the remaining amounts of the ATR mRNA were assessed using qRT-PCR. The values at time 0 were considered as 100%. The dashed lines reveal the half-life (50% mRNA remaining). Error bars indicate mean ± SD (n = 3). b RNAs bound to the AUF1 protein were immunoprecipitated from the indicated cells using anti-AUF1 antibody or anti-IgG (used as control), and then the mRNAs of the indicated genes were amplified by qRT-PCR. Data were normalized to the levels of the highly abundant GAPDH mRNA in each IP sample and represented as the enrichment of each mRNA. Error bars indicate mean ± SD. c Scheme showing ATR 3′UTR constructs used in this study. d HeLa and HEK293 cells were co-transfected with the nanoluciferase (NanoLuc) reporters bearing the indicated constructs, and control firefly luciferase expression vector for 16 h. Cells were lysed and luciferase activity was quantitated as Nanoluc/Firefly luc intensity ratio. e CAF-64 cells expressing AUF1 ORF (p37AUF1) or not (control) were co-transfected with nanoluciferase (NanoLuc) reporters bearing the indicated 3′UTRs, and control firefly luciferase expression vector for 16 h. Cells were lysed and luciferase activity was quantitated as Nanoluc/Firefly luc intensity ratio. Data in d and e are mean ± SEM of triplicate readings of three experiments for each cell line (**P < 0.0005, ***P = 0.0001). f Schematic representation of IL-6-dependent downregulation of ATR through STAT3/AUF1-related destabilization of the ATR mRNA