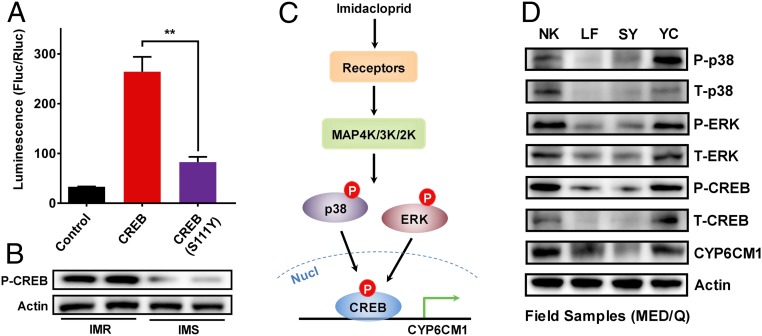
Fig. 4.
Model of the regulation of CYP6CM1 in B. tabaci. (A) Expression driven by the promoter of CYP6CM1 in the presence of wild type or mutated (Ser111Tyr) CREB. pGL4.10-CYP6CM1−939 to +1 and pGL4.73 plasmids were cotransfected into S2 cells with either CREB or mutated CREB (Ser111Tyr) or the pAC5.1b empty vector (control) and expression assessed using dual luciferase reporter assays (n = 3, mean ± SE; **P < 0.01, two-tailed Student’s t test). (B) Phosphorylation levels of Ser111 in CREB in the IMR and IMS strain as assessed by Western blot. Actin was used as a loading control. (C) A model of the regulation of CYP6CM1 in B. tabaci. In this model, exposure to imidacloprid is detected by an unknown receptor that actives mitogen-activated protein kinases of the MAPK pathway to phosphorylate ERK and p38. These, in turn, phosphorylate CREB, which then activates the expression of CYP6CM1. Activation and overexpression of CREB in the resistant strain increases the production of CYP6CM1 protein, which reduces the toxicity of imidacloprid. (D) Levels of Total-CYP6CM1, Total-CREB, Total-ERK, Total-p38, and phosphorylated CREB, ERK, and p38 in four field strains of B. tabaci MED. Actin was used as a loading control.