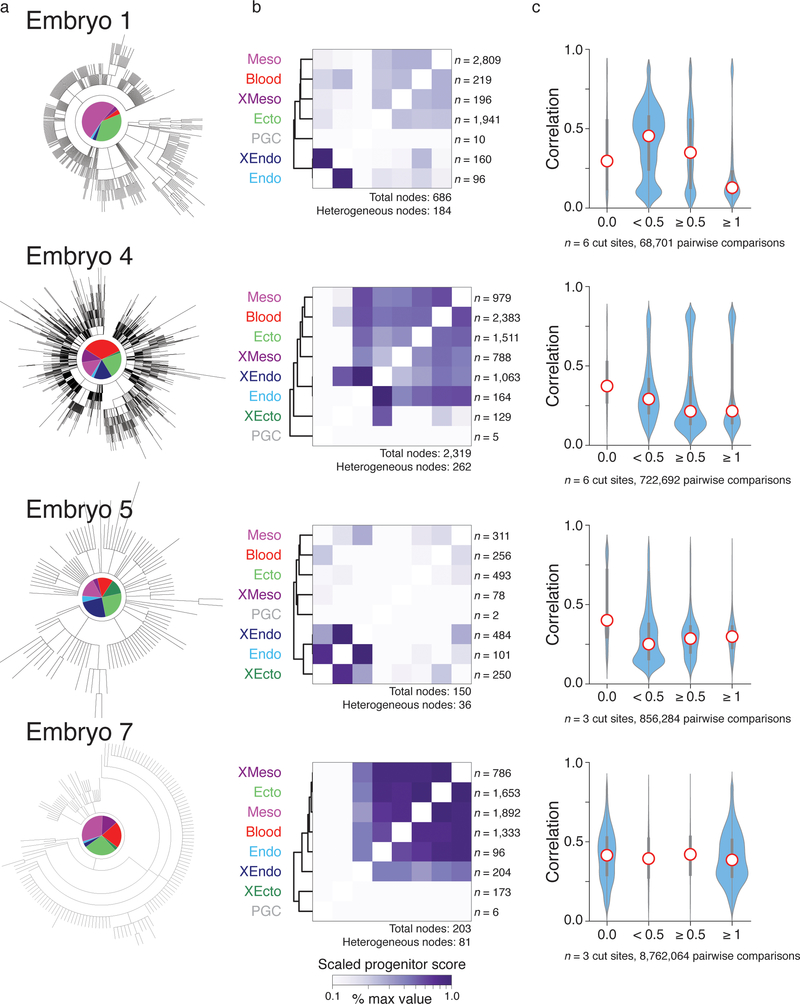
Extended Data Figure 9: Summary of results from additional mouse embryos.
Representative highest likelihood tree analyses for additional embryos, including:
a. Reconstructed trees as shown in Figure 4a.
b. Shared progenitor score heatmaps as shown in Figure 5a, normalized to the highest score for each embryo to account for differences in total node numbers. Here, the shared progenitor score is calculated as the number of nodes that are shared between tissues scaled by the number of number of tissues within each node (a shared score is calculated as 2−(n−1) where n is the number of clusters present within that node). In general, clustering of shared progenitors is recapitulated across embryos, with mesoderm and ectoderm sharing the highest relationship and either extra-embryonic ectoderm or extra-embryonic endoderm representing the most deeply rooted and distinct outgroup, though these scores are sensitive to the number of target sites and the rate of cutting. By shared progenitor, PGCs are also frequently distant from other embryonic tissues, but this often reflects the rarity of these cells, which restricts them to only a few branches of the tree in comparison to more represented germ layers. The number of heterogeneous nodes from which scores are derived is included for each heatmap.
c. Violin plots representing the pairwise relationship between lineage distance and transcriptional profile as shown for embryo 2 in Figure 4c. Lineage distance is calculated using a modified Hamming distance and transcriptional similarity by Pearson correlation. The exact dynamic range for lineage distance depends on the number of intBCs included and the cutting rate of the three guide array. Here, distances are binned into perfect (0), close (0 > x >0.5), intermediate ( 0.5 ≤ x < 1), and distant (x ≥ 1) relationships for all cells containing either 3 or 6 cut sites, depending on the embryo. As lineage distance increases, transcriptional similarity decreases, consistent with functional restriction over development. Red dot highlights the median, edges the interquartile range, and whiskers the full range.