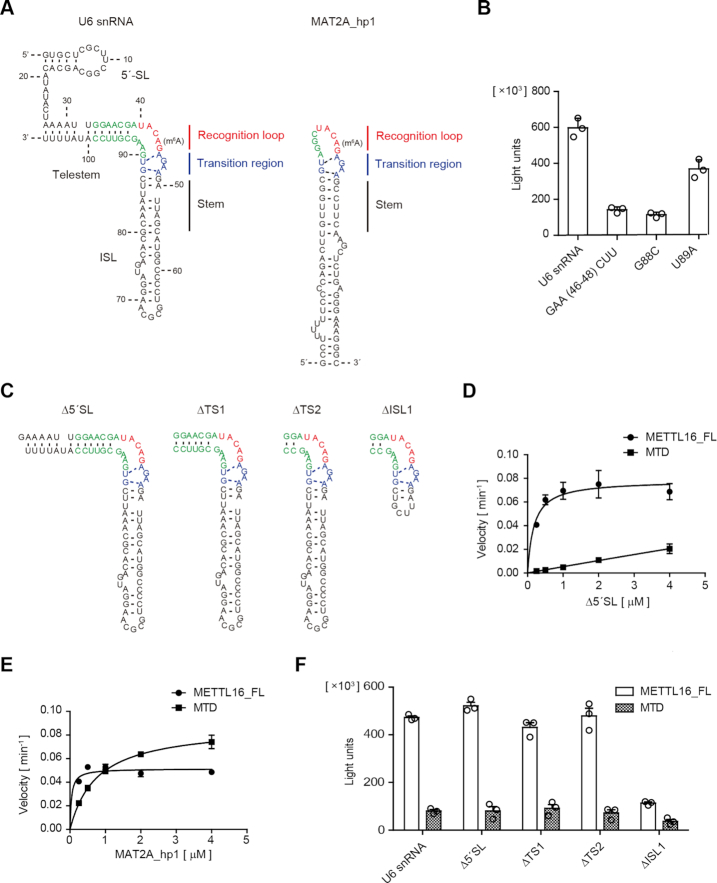
Figure 4.
Secondary structure of U6 snRNA for productive catalysis. (A) The secondary structural model of U6 snRNA for methylation by METTL16 (left) is similar to the structure of the MAT2A hairpin (MAT2A_hp1) in the complex with the MTD (right). The MAT2A_hp1 consists of the recognition loop, transition region, and stem required for productive catalysis after complex formation with the MTD. (B) The methylation of U6 snRNA variants with a mutation in the putative transition region, under the standard conditions (1 μM of RNAs). (C) Secondary structures of the U6 snRNA variants used for the assays in (D) and (F). (D) Steady-state kinetics of m6A modification of U6 snRNA_Δ5′SL by METTL16_FL and MTD. (E) Steady-state kinetics of m6A modification of MAT2A_hp1 by METTL16_FL and MTD (Supplementary Figure S1D). (F) The methylation of U6 snRNA variants by METTL16_FL and MTD under the standard conditions (1 μM of RNAs). Bars in the graphs in (B) and (D)–(F) are standard deviations (SD) of three independent experiments.